2024
- S. Jang, C. M. Schroeder, C. M. Evans, “Multiple Energy Dissipation Modes in Dynamic Polymer Networks with Neutral and Ionic Junctions”, Chemical Communications, in press (2024).
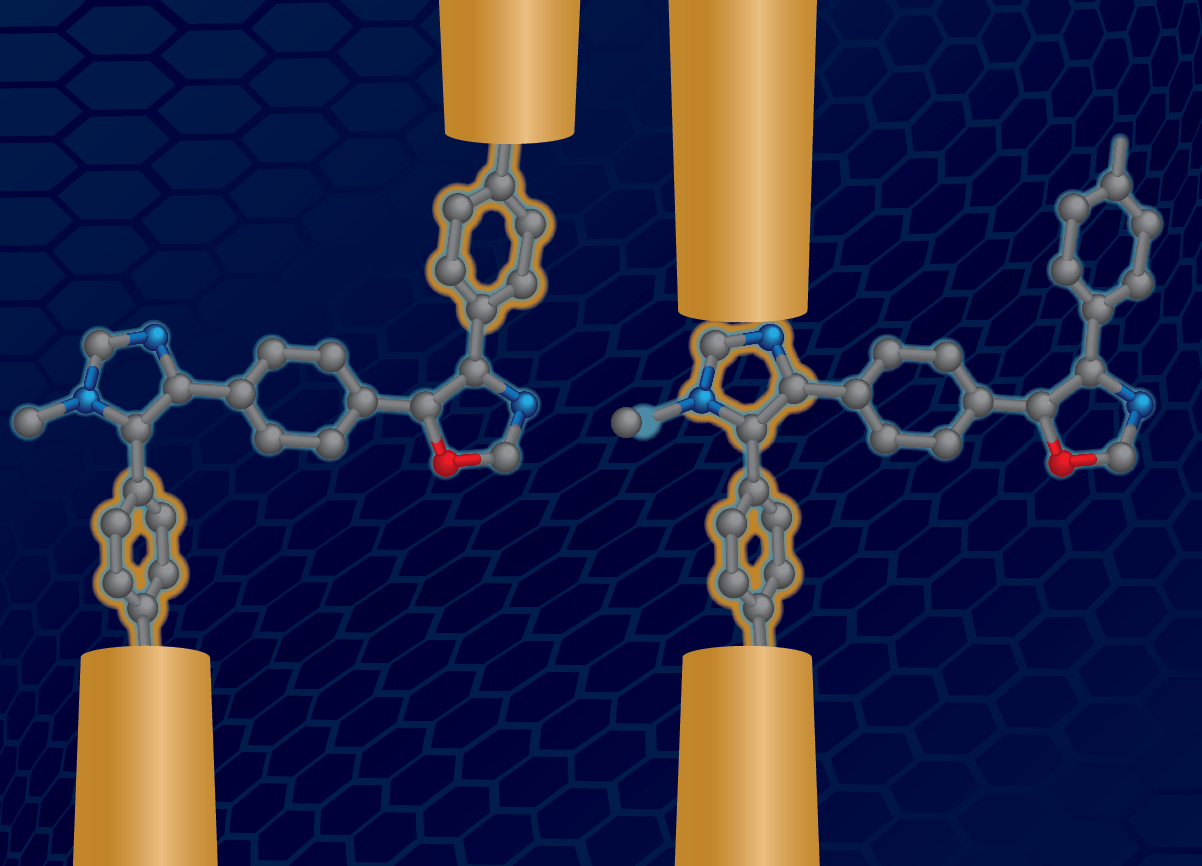
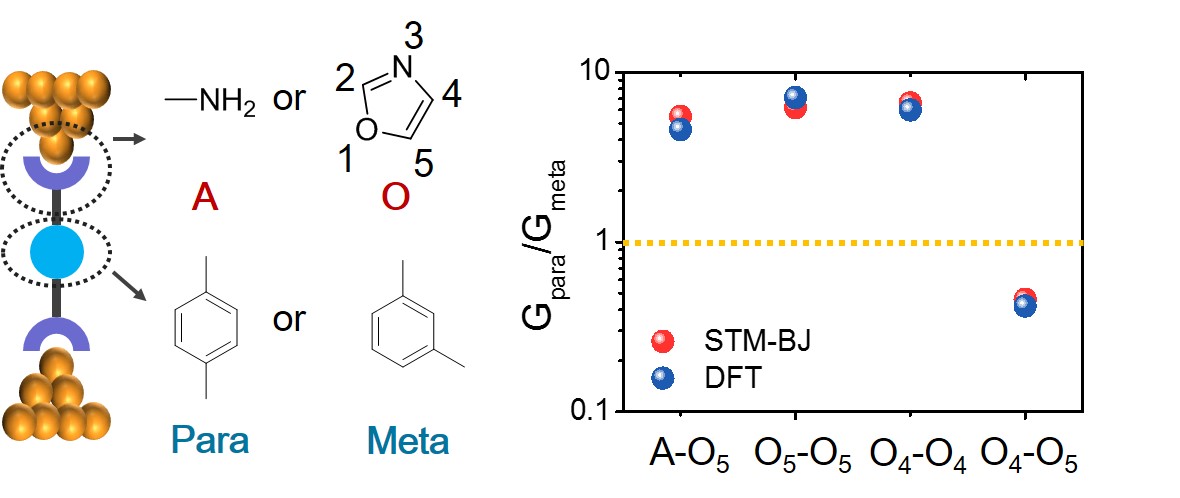
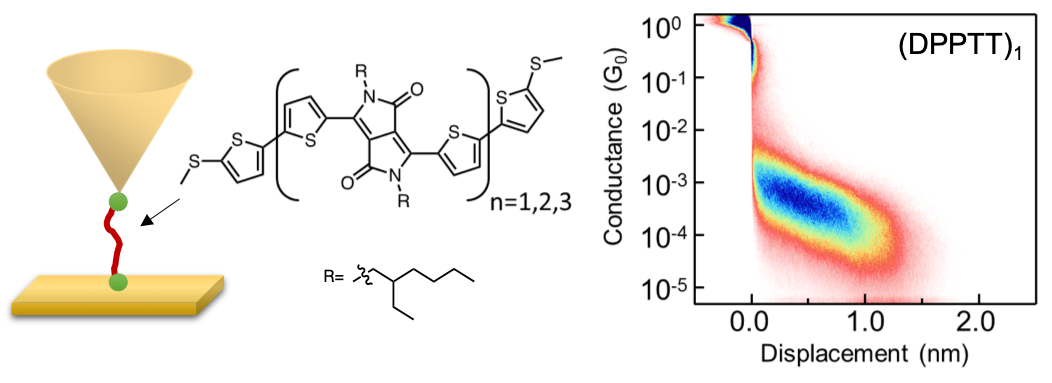
- X. Liu, H. Yang, H. Harb, R. Samajdar, T. J. Woods, O. Lin, Q. Chen, A. I. B. Romo, J. Rodríguez-López, R. S. Assary, J. S. Moore, C. M. Schroeder, “Shape-persistent Molecules Exhibit Nanogap-independent Conductance in Single-molecule Junctions”, Nature Chemistry, in press (2024). ChemRxiv link
- N. H. Angello, D. M. Friday, C. Hwang, S. Yi, A. H. Cheng, T. C. Torres Flores, E. R. Jira, W. Wang, A. Aspuru-Guzik, M. D. Burke*, C. M. Schroeder*, Y. Diao*, N. E. Jackson*, “Closed-Loop Transfer Enables AI to Yield Chemical Knowledge”, Nature, in press (2024). ChemRxiv link
- R. Samajdar, M. Meigooni, H. Yang, J. Li, M. A. Mosquera, N. E. Jackson, E. Tajkhorshid, C. M. Schroeder*, “Secondary Structure Determines Electron Transport in Peptides”, Proceedings of the National Academy of Sciences, 121, e2403324121 (2024). Web PDF
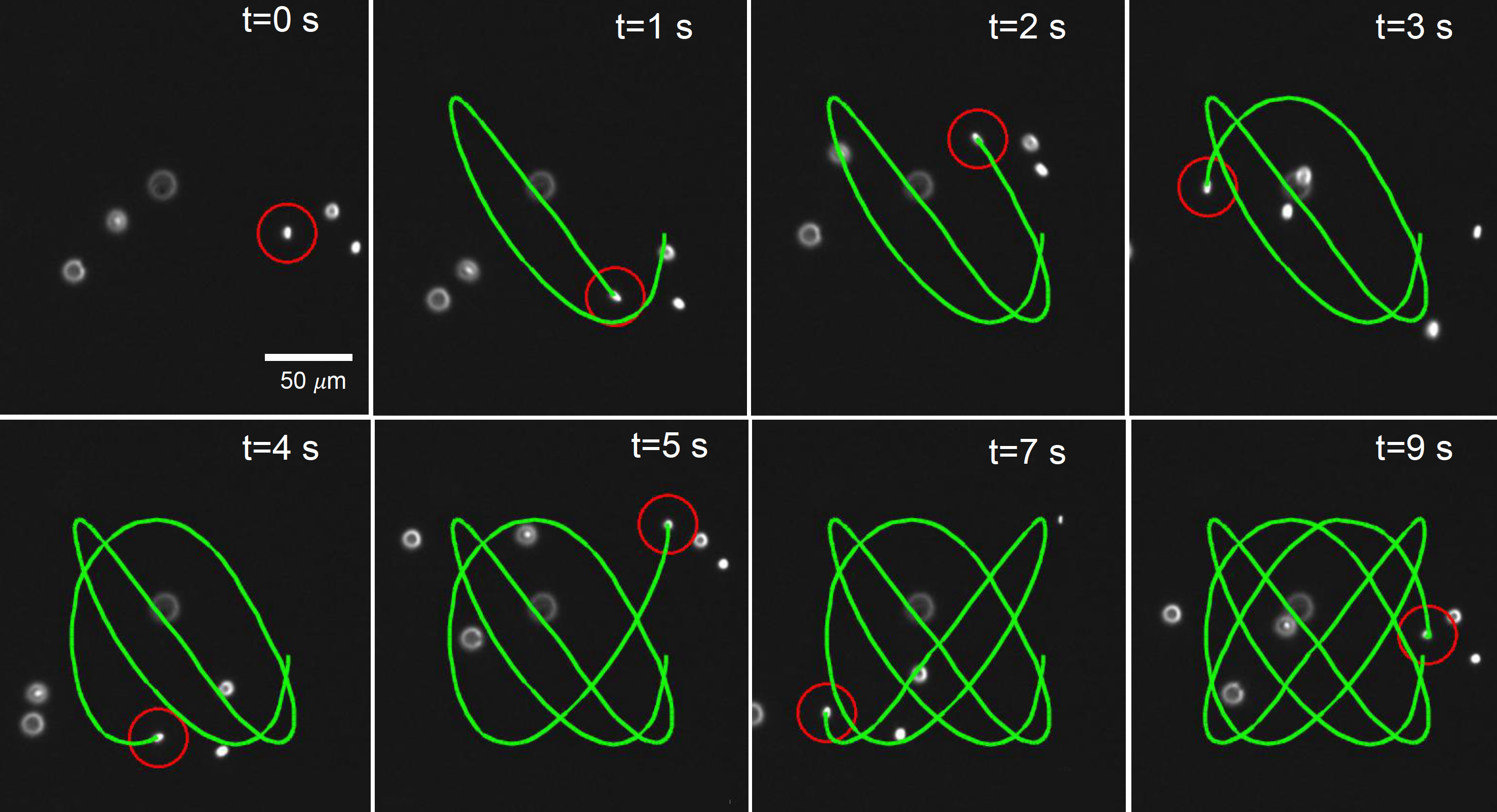
- S. Chaudhary, S. S. Velankar, C. M. Schroeder, “Dynamics of Meniscus-Bound Particle Clusters in Extensional Flow”, Journal of Rheology, 68, 397-413 (2024). Featured on Front Cover. Web PDF
- F. Kalutantirige, J. He, L. Yao, S. Cotty, S. Zhou, J. Smith, E. Tajkhorshid, C. M. Schroeder, J. S. Moore, H. An, X. Su, Y. Li, Q. Chen, “Beyond Nothingness: Formation and Functional Relevance of Voids in Polymer Films”, Nature Communications, 15, 2852 (2024). Web (open access)
- S. Kim and C. M. Schroeder, “Scaling Up Discovery”, Nature Synthesis, (2024). DOI: 10.1038/s44160-024-00498-5. News and Views article. Web PDF
2023
- I. Oh, M. Pence, N. Lukhanin, O. Rodriguez, C. M. Schroeder*, J. Rodriguez-Lopez*, “The Electrolab: An Open-Source, Modular Electrochemical Platform for Automated Characterization of Redox-Active Electrolytes”, Device, 1, 100103 (2023). Web PDF
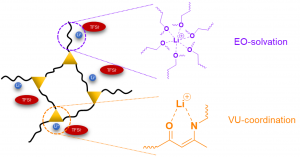
- S. Jang, E. Hernandez Alvarez, B. Jing, C. Shen, A. Schleife, C. M. Schroeder*, C. Evans*, “Control of Lithium Salt Partitioning, Coordination, and Solvation in Vitrimer Electrolytes”, Chemistry of Materials, 35, 8039-8049 (2023). Web PDF
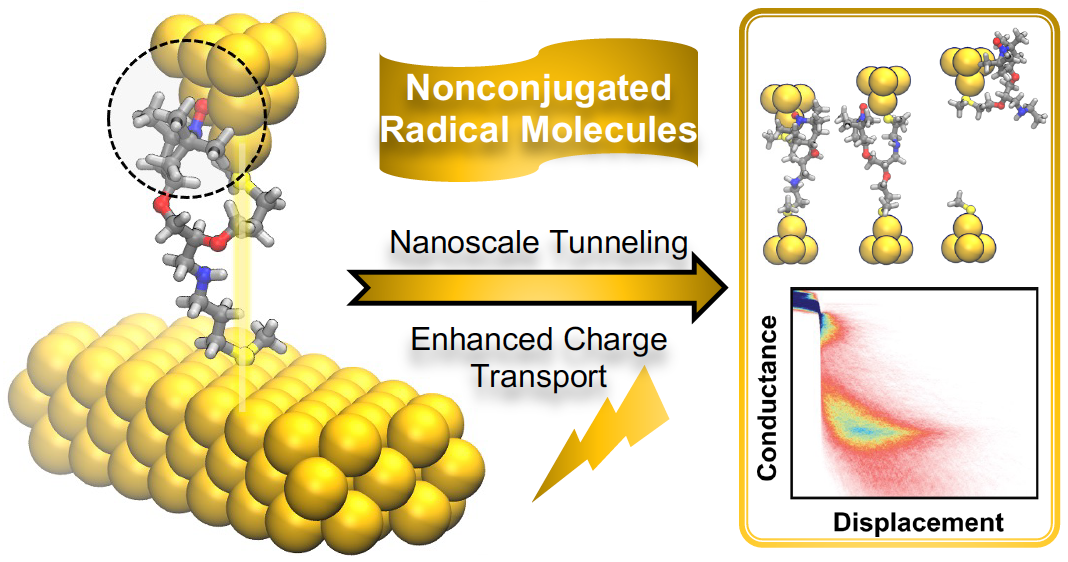
- Y. Tan*, J. Li*, S. Li, H. Yang, T. Chi, S. B. Shiring, K. Liu, B. M. Savoie, B. W. Boudouris*, C. M. Schroeder*, “Enhanced Electron Transport in Nonconjugated Radical Oligomers Occurs by Tunneling”, Nano Letters, 23, 5951-5958 (2023). Web PDF
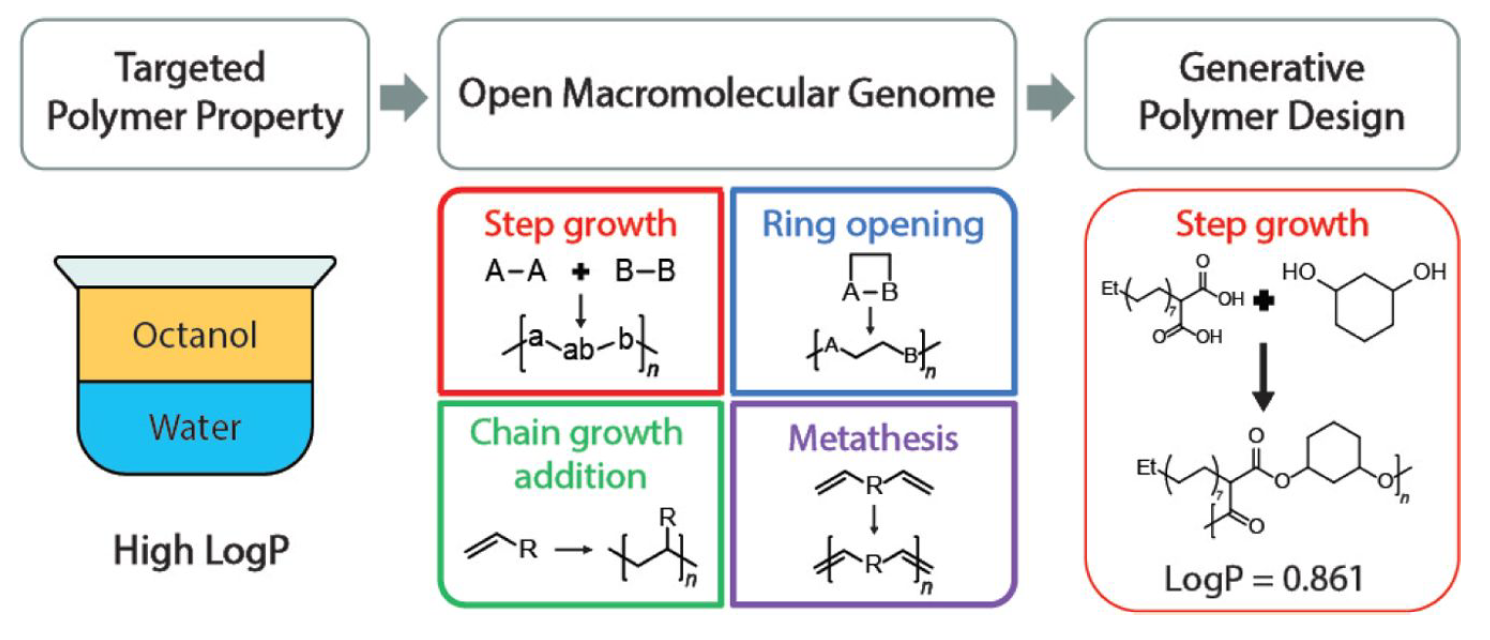
- S. Kim, C. M. Schroeder, N. E. Jackson, “Open Macromolecular Genome: Generative Design of Synthetically Accessible Polymers”, ACS Polymers Au, 3, 318-330 (2023). Web (open access)
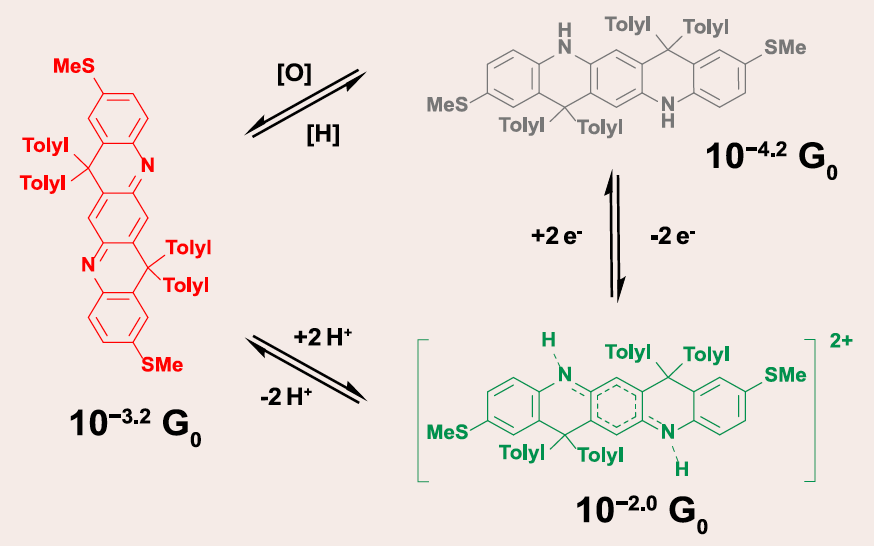
- J. Li*, B.-J. Peng*, S. Li, D. P. Tabor, L. Fang*, C. M. Schroeder*, “Ladder-type Conjugated Molecules as Robust Multi-state Single-Molecule Switches”, Chem, 9, 1-16 (2023). Web PDF
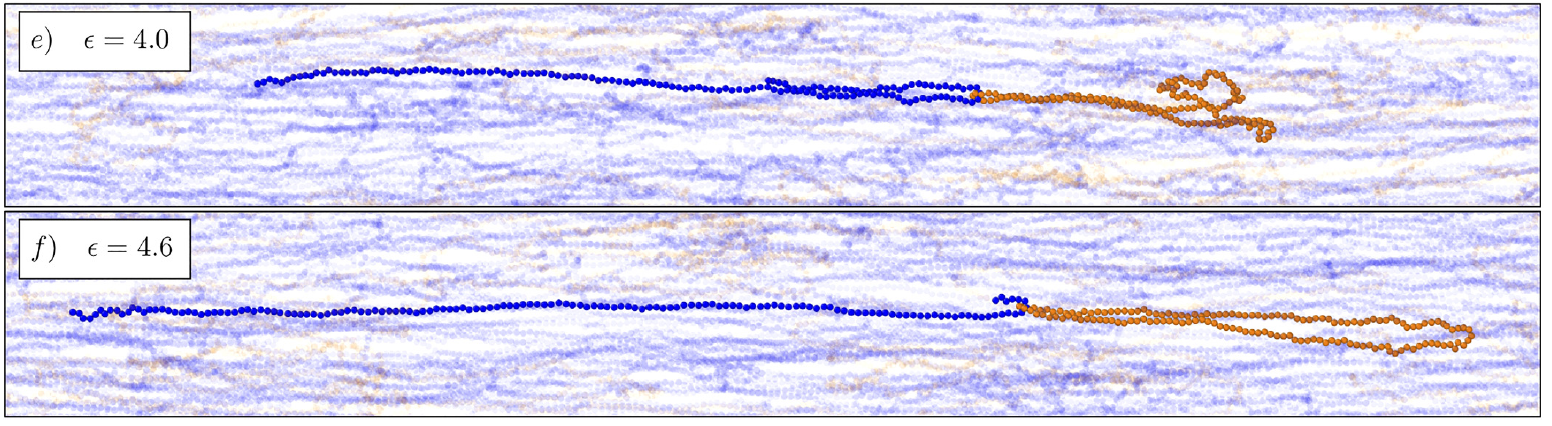
- M. Q. Tu, O. Davydovich, B. Mei, P. K. Singh, G. S. Grest, K. S. Schweizer, T. C. O’Connor, C. M. Schroeder, “Unexpected Slow Relaxation Dynamics in Pure Ring Polymers Arise from Intermolecular Interactions”, ACS Polymers Au, 3, 307-317 (2023). Web PDF
- M. Q. Tu*, H. V. Nguyen*, E. Foley, M. I. Jacobs, C. M. Schroeder, “3D Manipulation and Dynamics of Soft Materials in 3D Flows”, Journal of Rheology, 67, 877-890 (2023). Web PDF
- P. Banerjee, G. R. Burks, S. B. Bialik, M. Nassr, E. Bello, M. Alleyne, B. D. Freeman, J. E. Barrick, C. M. Schroeder, D. J. Milliron, “Nanostructure-derived Anti-reflectivity in Leafhopper Brochosomes”, Advanced Photonics Research, 4, 2200343 (2023). Web PDF
- M. Pence, O. Rodríguez, N. Lukhanin, C. M. Schroeder, J. Rodríguez-López, “Automated Measurement of Electrogenerated Redox Species Degradation Using Multiplexed Interdigitated Electrode Arrays”, ACS Measurement Science Au, 3, 62-72 (2023). Web PDF
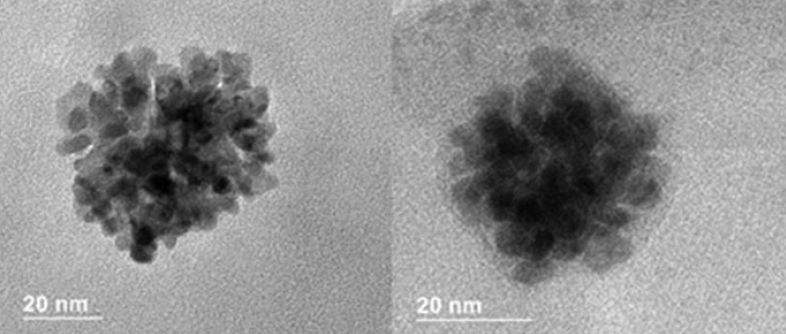
- G. R. Burks, L. Yao, F. Kalutantirige, K. Gray, E. Bello, S. Rajagopalan, S. Bialik, J. Barrick, M. Alleyne, Q. Chen, C. M. Schroeder, “Electron Tomography and Machine Learning for Understanding the Highly Ordered Structure of Leafhopper Brochosomes”, Biomacromolecules, 24, 190-200 (2023). Web PDF
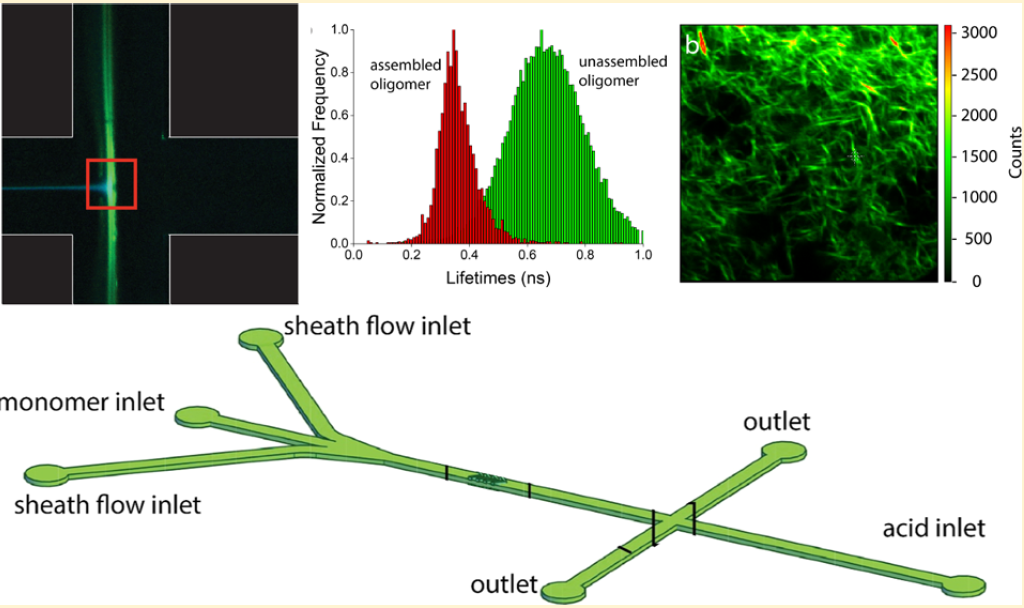
- S. Qu, Zihao Ou, Y. Savsatli, L. Yao, Y. Cao, E. C. Montoto, H. Yu, J. Hui, B. Li, J. A. N. T. Soares, L. Kisley, B. Bailey, E. A. Murphy, J. Liu, C. M. Evans, C. M. Schroeder, J. Rodríguez-López, J. S. Moore, Q. Chen, P. V. Braun, “Visualizing Energy Transfer Between Redox-Active Colloids”, submitted (2023). arXiv link
2022
- H. Yu, F. Kalutantirige, L. Yao, C. M. Schroeder*, Q. Chen*, J. S. Moore*, “Self-Assembly of Repetitive Segment and Random Segment Polymer Architectures”, ACS Macro Letters, 11, 1366-1372 (2022). Web PDF
- N. H. Angello, V. Rathore, W. Beker, A. Wolos, E. R. Jira, R. Roszak, T. C. Wu, C. M. Schroeder, A. Aspuru-Guzik, B. A. Grzybowski, M. D. Burke, “Closed-loop Optimization of General Reaction Conditions”, Science, 378, 399-405 (2022). Web PDF
News coverage: C&E News, Phys Org, EurekAlerts, ScienMag, Nanotechnology Now, Bioengineer.org, Nanowerk
- M. I. Jacobs, P. Bansal, D. Shukla, C. M. Schroeder, “Understanding Supramolecular Assembly of Supercharged Proteins”, ACS Central Science, 8, 1350–1361 (2022). Web PDF
- P. Singh, M. Pacholski, J. Gu, Y.-K. Go, G. Singhal, C. Leal, P. Braun, K. Patankar, R. Drumright, S. A. Rogers, C. M. Schroeder, “Designing Multicomponent Polymer Colloids for Self-stratifying Films”, Langmuir, 38, 11160-11170 (2022). Web PDF
- P. Kafle, S. Huang, K. S. Park, F. Zhang, H. Yu, C. Kasprzak, H. Kim, C. M. Schroeder, A. van der Zande, Y. Diao, “Role of Interfacial Interactions in Graphene-Directed Assembly of Monolayer Conjugated Polymers”, Langmuir, 38, 6984-6995 (2022). Web PDF
- H. Yu*, J. Li*, S. Li, Y. Liu, N. E. Jackson, J. S. Moore, C. M. Schroeder, “Efficient Intermolecular Charge Transport in π-Stacked Pyridinium Dimers using Cucurbit[8]uril Supramolecular Complexes”, Journal of the American Chemical Society, 144, 3162-3173 (2022). Web PDF
- C. Pan*, S. K. Tabatabaei*, S. M. H. Tabatabaei Yazdi, A. G. Hernandez, C. M. Schroeder*, O. Milenkovic*, “Rewritable Two-Dimensional DNA-Based Data Storage with Machine Learning Reconstruction”, Nature Communications, 13, 2984 (2022). Web PDF
- S. Li, E. R. Jira, N. H. Angello, J. Li, H. Yu, J. S. Moore, Y. Diao, M. D. Burke, C. M. Schroeder, “Using Automated Synthesis to Understand the Role of Side Chains on Molecular Charge Transport”, Nature Communications, 13, 2101 (2022). Web PDF
News coverage: Phys Org, EurekAlerts, ScienMag, Nanotechnology Now, TVN, Bioengineer.org, Nanowerk
- S. K. Tabatabaei*, B. Pham*, C. Pan*, J. Liu, S. Chandak, S. A. Shorkey, A. G. Hernandez, A. Aksimentiev*, M. Chen*, C. M. Schroeder*, O. Milenkovic*, “Expanding the Molecular Alphabet of DNA-Based Data Storage Systems with Neural Network Nanopore Readout Processing”, Nano Letters, 22, 1905-1914 (2022). Web PDF
News coverage: CNET, Phys Org, EurekAlerts, Science Daily, Electronics Weekly, Nanotechnology Now, Tech Times, Tech Register, Bioengineer.org, Nanowerk
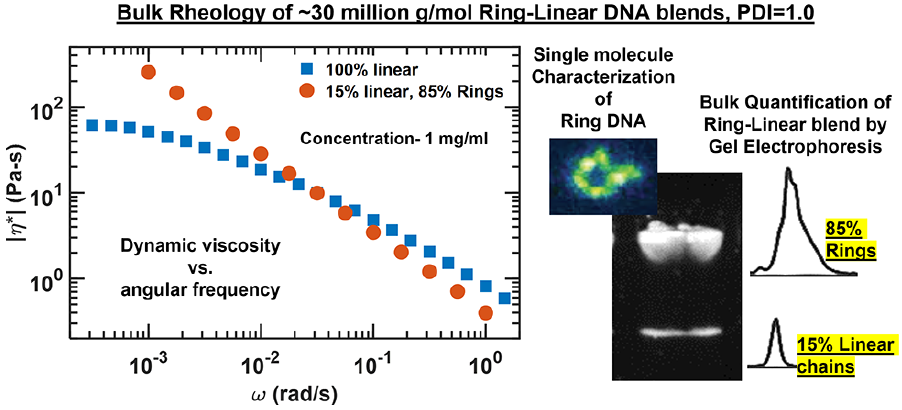
- D. Kong, S. Banik, M. J. San Francisco, M. Lee, R. M. Robertson-Anderson, C. M. Schroeder, G. B. McKenna, “Rheology of Entangled Solutions of Ring-linear DNA Blends”, Macromolecules, 55, 1205-1217 (2022). Web PDF
- A. Khandelwal, N. Athreya, M. Tu, L. Janavicius, Z. Yang, O. Milenkovic, J.-P. Leburton, C. M. Schroeder, X. Li, “Self-Assembled Microtubular Electrodes for On-Chip Low-Voltage Electrophoretic Manipulation of Charged Particles and Macromolecules”, Microsystems & Nanoengineering, 8, 22 (2022). Web PDF
- Y. Zhou, F. Latinwo, C. M. Schroeder, “Crooks Fluctuation Theorem for Single Polymer Dynamics in Time-dependent Flows: Understanding Viscoelastic Hysteresis”, Entropy, 24, 27 (2022). Web PDF
2021
- M. I. Jacobs, E. R. Jira, C. M. Schroeder, “Understanding How Coacervates Drive Reversible Small Molecule Reactions to Promote Molecular Complexity”, Langmuir, 37, 14323-14335 (2021). Web PDF
- D. Kumar and C. M. Schroeder, “Nonlinear Transient and Steady-state Stretching of Deflated Vesicles in Flow”, Langmuir, 37, 13976-13984 (2021). Web PDF
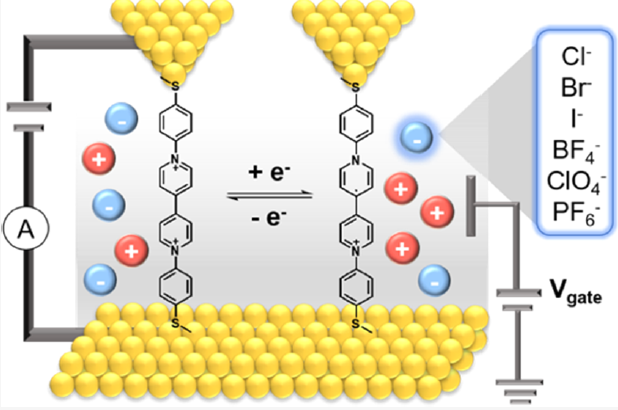
- J. Li, S. Pudar, H. Yu, S. Li, J. S. Moore, J. Rodríguez-López, N. E. Jackson, C. M. Schroeder, “Reversible Switching of Molecular Conductance in Viologens is Controlled by the Electrochemical Environment”, Journal of Physical Chemistry C, 125, 21862-21872 (2021). Web PDF
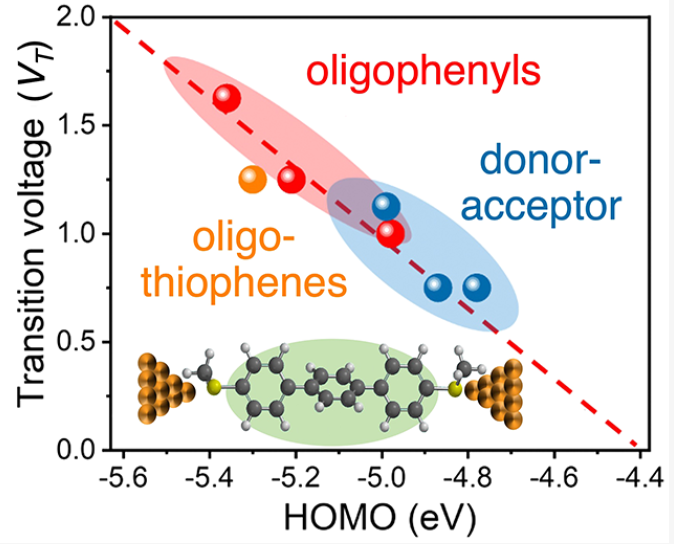
- S. Li, H. Yu, J. Li, N. Angello, E. R. Jira, B. Li, M. D. Burke, J. S. Moore, C. M. Schroeder, “Transition between Non-resonant and Resonant Charge Transport in Molecular Junctions”, Nano Letters, 21, 8340-8347 (2021). Web PDF
- C. Lin, D. Kumar, C. Richter, S. Wang, C. M. Schroeder*, V. Narsimhan*, “Vesicle Dynamics in Large Amplitude Oscillatory Extensional Flow”, Journal of Fluid Mechanics, 929 (2021). Web PDF
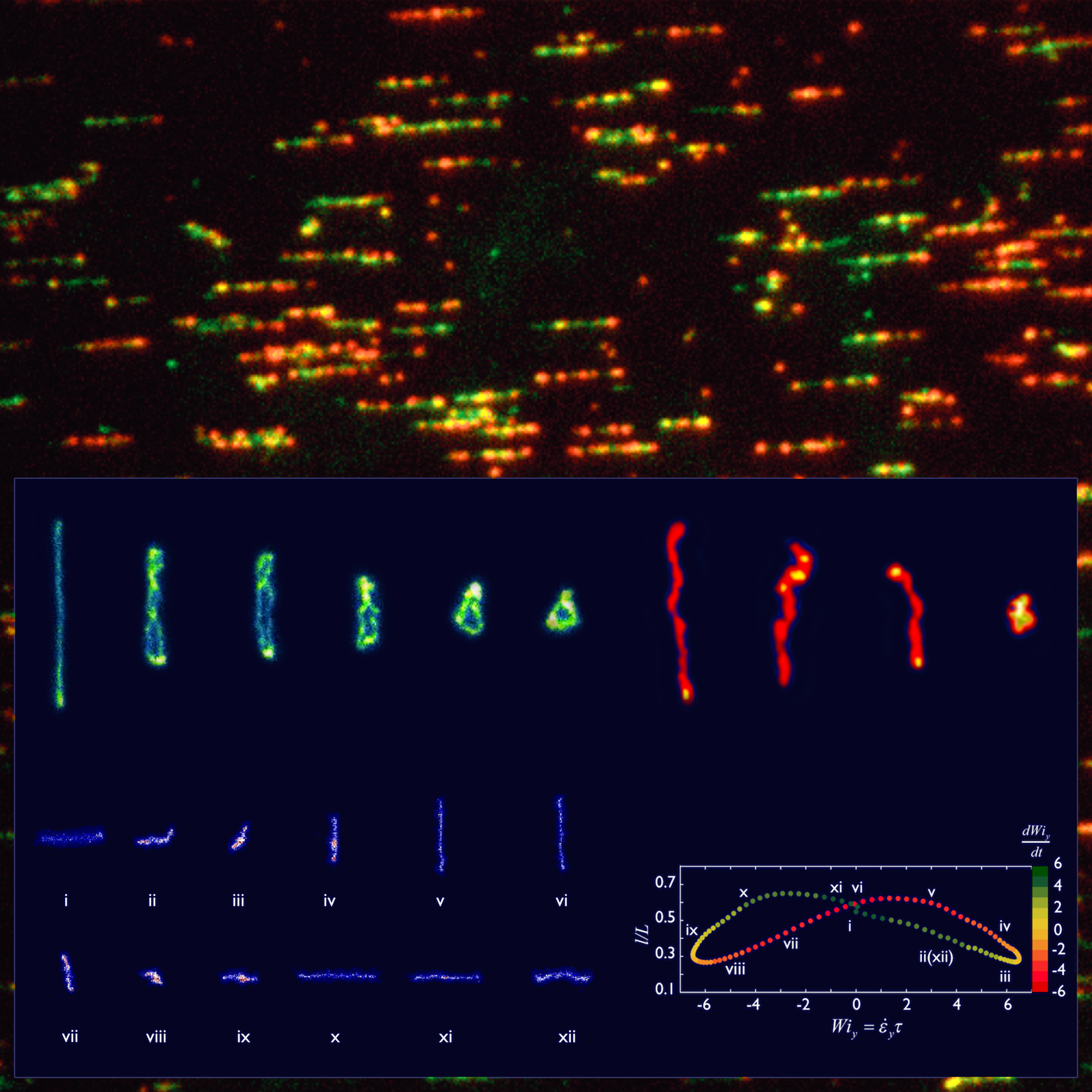
- Y. Zhou, C. D. Young, K. E. Regan, M. Lee, S. Banik, D. Kong, G. B. McKenna, R. M. Robertson-Anderson, C. E. Sing, C. M. Schroeder, “Dynamics and Rheology of Ring-Linear Blend Semidilute Solutions in Extensional Flow: Single Molecule Experiments”, Journal of Rheology, 65, 729-744 (2021). Web PDF
- C. D. Young, Y. Zhou, C. M. Schroeder, C. E. Sing, “Dynamics and Rheology of Ring-Linear Blend Semidilute Solutions in Extensional Flow: Modeling and Molecular Simulations”, Journal of Rheology, 65, 757-777 (2021). Web PDF
- S. Jain, S. Shukla, C. Yang, M. Zhang, Z. Fatma, M. Lingamaneni, S. Abesteh, S. T. Lane, X. Xiong, Y. Wang, C. M. Schroeder, P. R. Selvin, H. Zhao, “TALEN Outperforms Cas9 in Editing Heterochromatin Target Sites”, Nature Communications, 12, 606 (2021). Web PDF
News coverage: Phys Org, EurekAlert, ScienMag, Science Daily, The Medical News, Bioengineer.org
2020
- S. Patel, C. D. Young, C. E. Sing, C. M. Schroeder, “Non-monotonic Dependence of Comb Polymer Relaxation on Branch Density in Semi-dilute Solutions”, Physical Review Fluids, 5, 121301R (2020). Web PDF
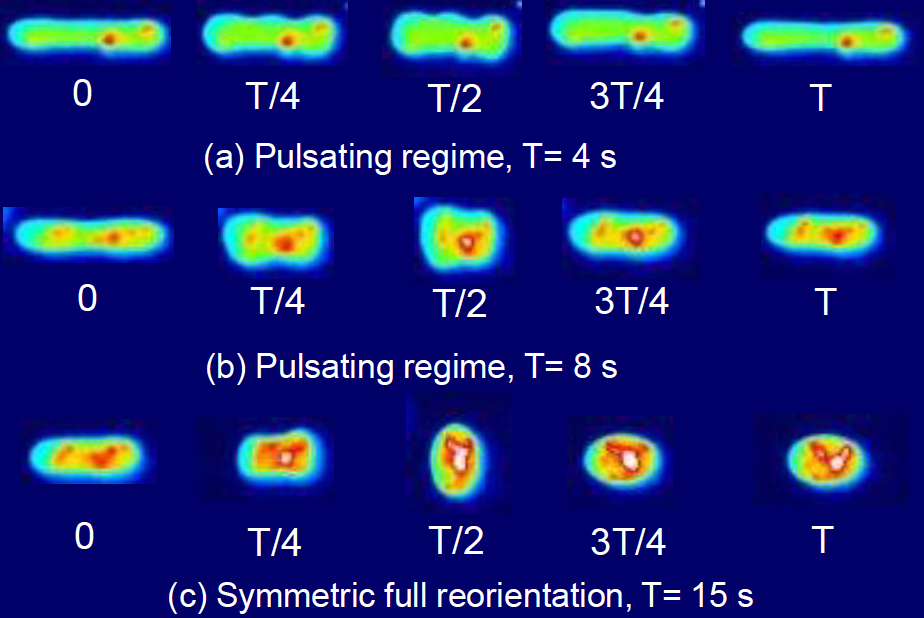
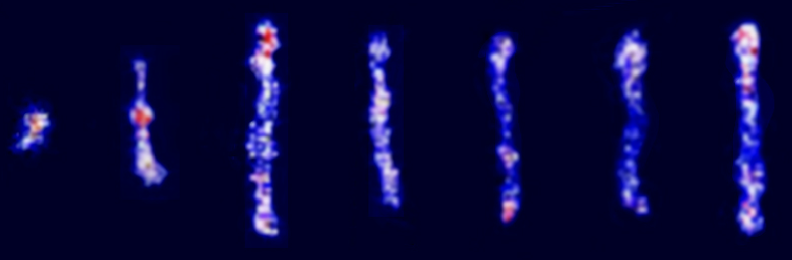
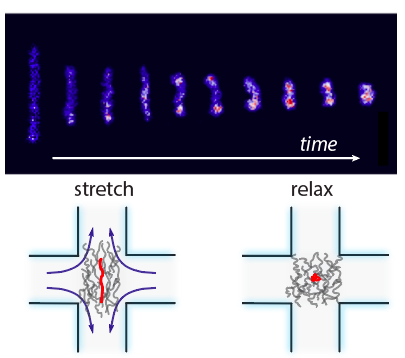
- M. Tu, M. Lee, R. M. Robertson-Anderson, C. M. Schroeder, “Direct Observation of Ring Polymer Dynamics in the Flow-Gradient Plane of Shear Flow”, Macromolecules, 53, 9406–9419 (2020). Web PDF
- D. Mai and C. M. Schroeder, “100th Anniversary of Macromolecular Science Viewpoint: Single-Molecule Studies of Synthetic Polymers”, ACS Macro Letters, 9, 1332-1341, (2020). Web PDF
- D. Kumar, C. M. Richter, C. M. Schroeder, “Double-mode Relaxation of Highly Deformed Vesicles”, Physical Review E, 102, 010605R (2020). Web PDF
- S. Li, H. Yu, X. Chen, A. A. Gewirth, J. S. Moore, C. M. Schroeder, “Covalent Ag-C Bonding Contacts from Unprotected Terminal Acetylenes for Molecular Junctions”, Nano Letters, 20, 5490–5495 (2020). Web PDF
- K. R. Peddireddy, M. Lee, C. M. Schroeder, R. M. Robertson-Anderson, “Viscoelastic Properties of Ring-linear DNA Blends Exhibit Non-monotonic Dependence on Blend Composition”, Physical Review Research, 2, 023213 (2020). Web PDF
- E. R. Jira, K. Shmilovich, T. S. Kale, A. Ferguson, J. D. Tovar, C. M. Schroeder, “Effect of Core Oligomer Length on the Phase Behavior and Assembly of pi-conjugated Peptides”, ACS Applied Materials & Interfaces, 12, 20722-20732 (2020). Web PDF
- S. Li, J. Li, H. Yu, S. Pudar, B. Li, J. Rodríguez-Lopez, J. S. Moore, C. M. Schroeder, “Characterizing Intermolecular Interactions in Redox-active Pyridinium-based Molecular Junctions”, Journal of Electroanalytical Chemistry, 85, 114070 (2020). Web PDF
- D. Kumar, A. Shenoy, J. C. Deutsch, C. M. Schroeder, “Automation and Flow Control for Particle Manipulation”, Current Opinion in Chemical Engineering, 29, 1-8 (2020). Web PDF
- H. Yu*, S. Li*, K. E. Schwieter, Y. Liu, B. Sun, J. S. Moore, C. M. Schroeder, “Charge Transport in Sequence-defined Conjugated Oligomers”, Journal of the American Chemical Society, 142, 4852-4861 (2020). Web PDF
- D. Kumar, C. M. Richter, C. M. Schroeder, “Conformational Dynamics and Phase Behavior of Lipid Vesicles in a Precisely Controlled Extensional Flow”, Soft Matter, 16, 337-347 (2020). Web PDF
- K. Peddireddy, M. Lee, Y. Zhou, S. Adalbert, C. M. Schroeder, R. Robertson-Anderson, “Unexpected Entanglement Dynamics in Semidilute Blends of Supercoiled and Ring DNA”, Soft Matter, 16, 152-161 (2020). Web PDF
- L. Cuculis*, C. Zhao*, Z. Abil, H. Zhao, D. Shukla*, C. M. Schroeder*,”Divalent Cations Enhance TALE DNA-Binding Specificity”, Nucleic Acids Research, 48, 1406-1422 (2020). Web PDF
2019
- D. Kumar, A. Shenoy, S. Li, C. M. Schroeder, “Orientation Control and Nonlinear Trajectory Tracking of Colloidal Particles using Microfluidics”, Physical Review Fluids, 4, 114203 (2019). Web PDF
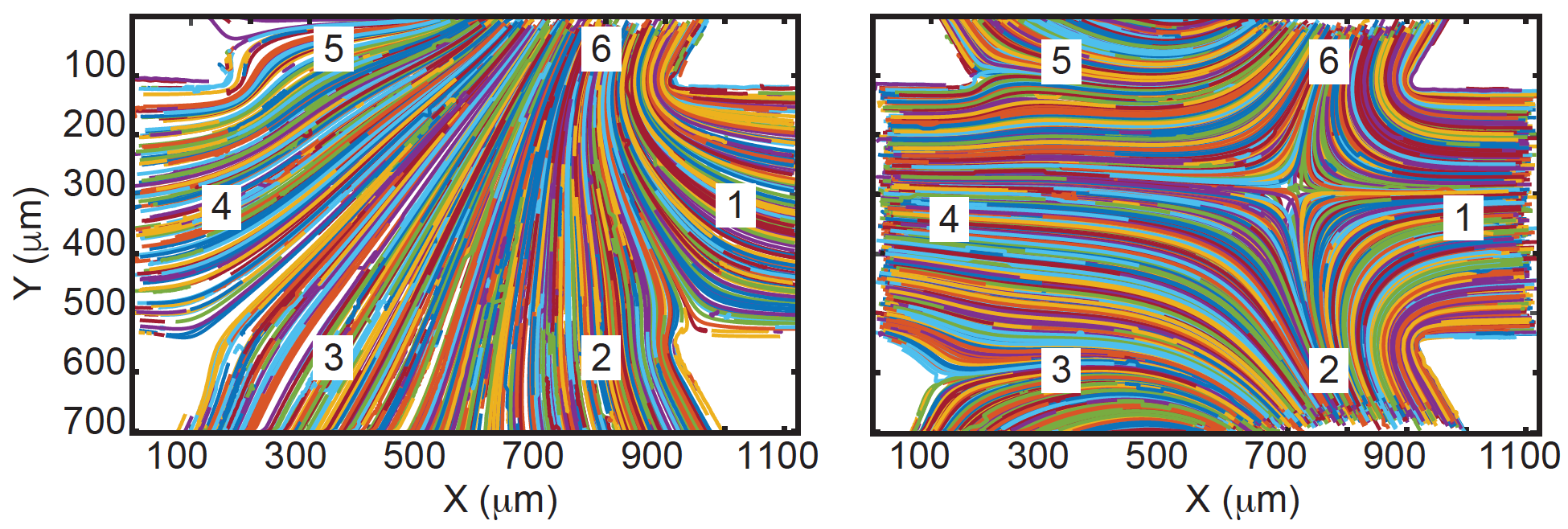
- A. Shenoy, D. Kumar, S. Hilgenfeldt, C. M. Schroeder, “Flow Topology During Multiplexed Particle Manipulation using a Stokes Trap”, Physical Review Applied, 12, 054010 (2019). Web PDF
- S. Li*, H. Yu*, K. E. Schwieter, K. Chen, B. Li, Y. Liu, J. S. Moore, C. M. Schroeder, “Charge Transport and Quantum Interference in Oxazole-Terminated Conjugated Oligomers”, Journal of the American Chemical Society, 141, 16079-16084 (2019). Web PDF
- L. Valverde, B. Li, C. M. Schroeder, W. Wilson, “In Situ Photophysical Characterization of pi-conjugated Oligopeptides Assembled via Continuous Flow Processing”, Langmuir, 35, 10947-10957 (2019). Web PDF
- C. Boucher-Jacobs, B. Li, C. M. Schroeder, and D. Guironnet, “Solubility and Activity of a Phosphinosulfonate Palladium Catalyst in Water with Different Surfactants”, Polymer Chemistry, 10, 1988-1992 (2019). Web PDF
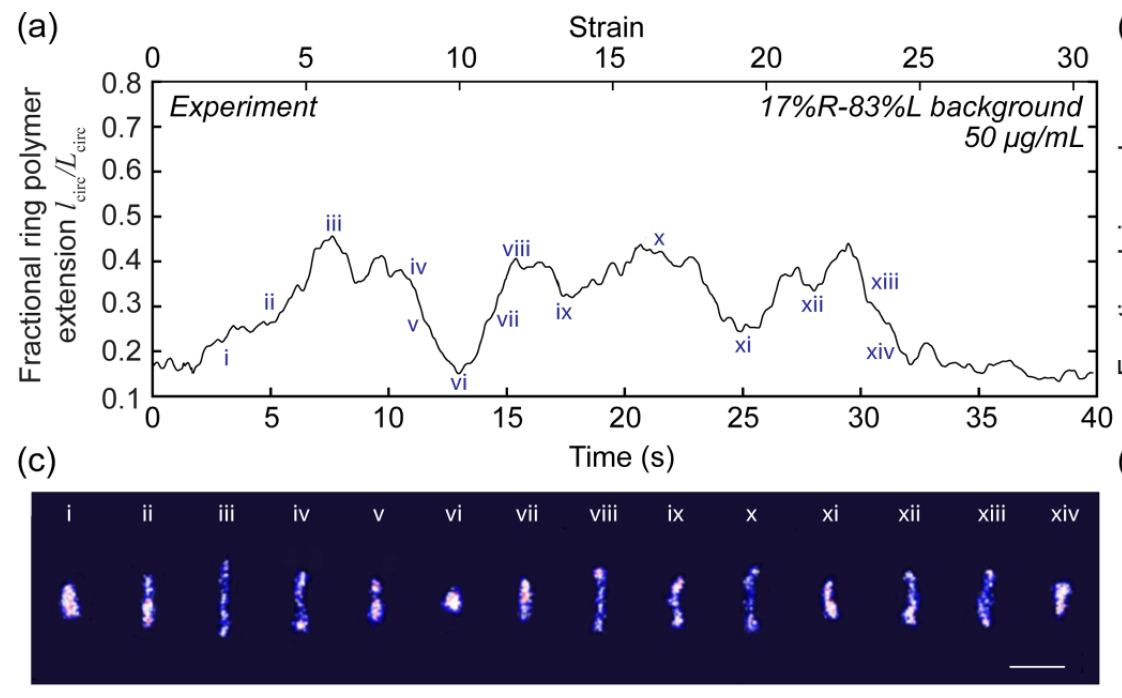
- Y. Zhou, K. W. Hsiao, K. E. Regan, D. Kong, G. B. McKenna, R. M. Robertson-Anderson, C. M. Schroeder, “Effect of Molecular Architecture on Ring Polymer Dynamics in Semidilute Linear Polymer Solutions”, Nature Communications, 10, 1753, DOI:10.1038/s41467-019-09627 (2019). Web PDF
- B. Li, H. Yu, E. C. Montoto, Y. Liu, S. Li, K. Schwieter, J. Rodriquez-Lopez, J. S. Moore, C. M. Schroeder, “Intrachain Charge Transport through Conjugated Donor-Acceptor Oligomers”, ACS Applied Electronic Materials, 1, 7-12, (2019). Web PDF
2018
- Y. Zhou and C. M. Schroeder, “Dynamically Heterogeneous Relaxation of Entangled Polymer Chains”, Physical Review Letters, 120, 267801 (2018). Web PDF
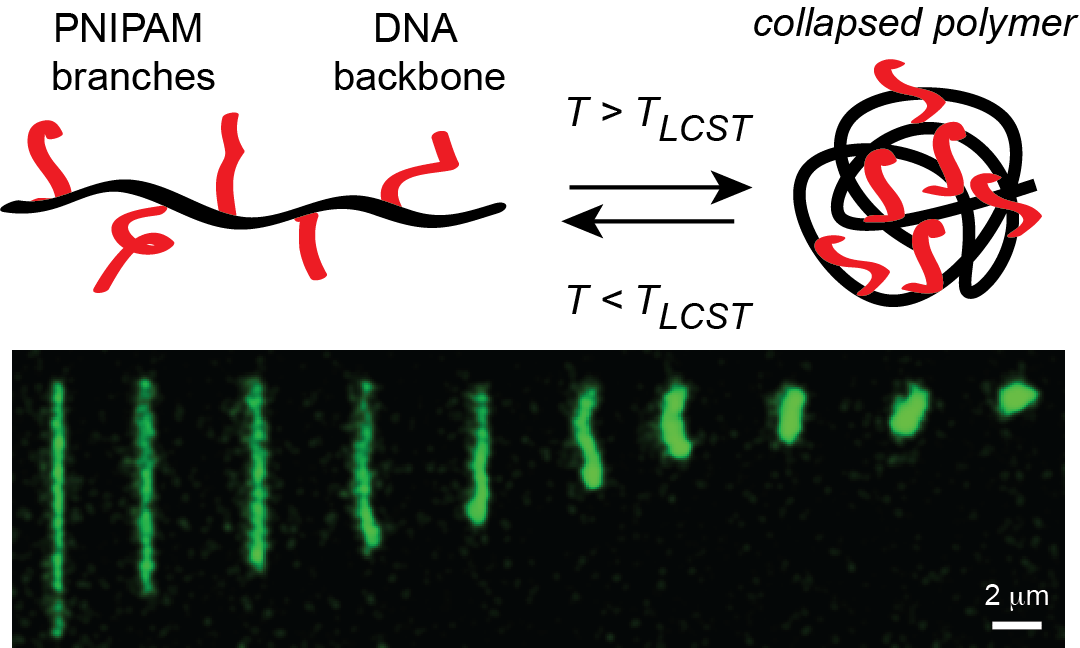
- S. Li and C. M. Schroeder, “Synthesis and Direct Observation of Thermo-Responsive DNA Copolymers”, ACS Macro Letters, 7, 281-286 (2018). Web PDF
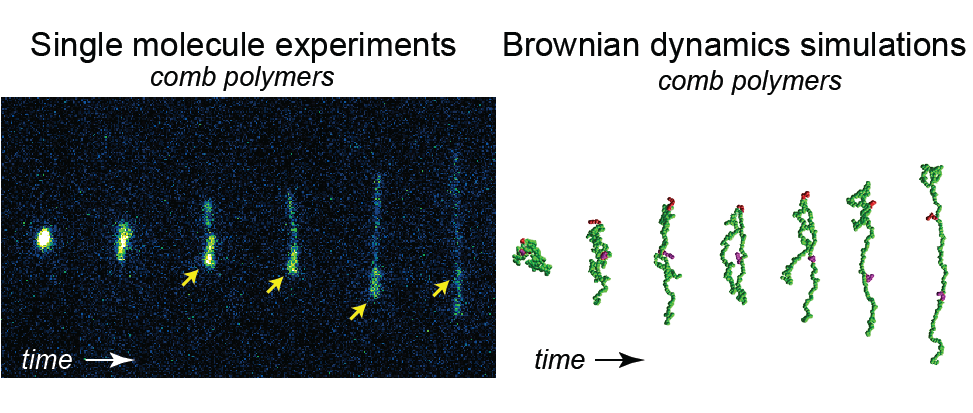
- D. J. Mai, A. Sadaat, B. Khomami, C. M. Schroeder, “Stretching Dynamics of Single Comb Polymers in Extensional Flow”, Macromolecules, 51, 1507-1517 (2018). Web PDF
- C. M. Schroeder, “Single Polymer Dynamics for Molecular Rheology”, Journal of Rheology, 62, 371-403 (2018). Web PDF
2017
- B. Li, L. R. Valverde, F. Zhang, Y. Zhou, S. Li, Y. Diao, W. L. Wilson, C. M. Schroeder, “Macroscopic Alignment and Assembly of pi-conjugated Oligopeptides using Colloidal Microchannels”, ACS Applied Materials & Interfaces, 9, 41586-41593 (2017). Web PDF
- S. Kumar, J. S. Katz, C. M. Schroeder, “Heterogeneous Drying and Non-monotonic Contact Angle Dynamics in Concentrated, Film-forming Latex Drops”, Physical Review Fluids, 2, 114304 (2017). Web PDF
- J. P. Berezney, A. B. Marciel, C. M. Schroeder, O. A. Saleh, “Scale-dependent Stiff ness and Internal Tension of a Model Brush Polymer”, Physical Review Letters, 116, 127801 (2017). Web PDF
- K. W. Hsiao, J. Dinic, Y. Ren, V. Sharma, C. M. Schroeder, “Passive Non-linear Microrheology for
Determining Extensional Viscosity”, Physics of Fluids, 29, 121603 (2017). Web PDF
- Y. Zhou, B. Li, S. Li, H. A. M. Ardoena, W. L. Wilson, J. D. Tovar, C. M. Schroeder, “Concentration-Driven Assembly and Sol-Gel Transition pi-Conjugated Oligopeptides”, ACS Central Science, DOI: 10.1021/acscentsci.7b00260 (2017). Web PDF
- L. W. Cuculis and C. M. Schroeder, “Molecular Mechanisms for Genome Editing Proteins: Single Molecule Studies of TALEs and CRISPR/Cas9”, Annual Review of Chemical and Biomolecular Engineering, 8, 577-597 (2017). Web PDF
- B. Li, S. Li, Y. Zhou, H. A. M. Ardona, L. R. Valverde, W. L. Wilson, J. D. Tovar, C. M. Schroeder, “Non-equilibrium Self-assembly of pi-conjugated Oligopeptides in Solution”, ACS Applied Materials & Interfaces, 9, 3977-3984 (2017). Web PDF
- C. Sasmal, K. Hsiao, C. M. Schroeder, J. R. Prakash, “Parameter-free Prediction of DNA Dynamics in Planar Extensional Flow of Semi-dilute Solutions”, Journal of Rheology, 61, 169-186 (2017). Web PDF
- K. Hsiao, C. Sasmal, J. R. Prakash, C. M. Schroeder, “Direct Observation of DNA Dynamics in Semi-dilute Solutions in Extensional Flow”, Journal of Rheology, 61, 151-167 (2017). Web PDF
2016
- Y. Zhou and C. M. Schroeder, “Transient and Average Unsteady Dynamics of Single Polymers in Large-amplitude Oscillatory Extension”, Macromolecules, 49, 8018-8030 (2016). Web PDF
- D. J. Mai and C. M. Schroeder, “Single Polymer Dynamics of Topologically Complex DNA”, Current Opinion in Colloid and Interface Science, 26, 28-40 (2016). Web PDF
- Y. Zhou and C. M. Schroeder, “Single Polymer Dynamics in Large Amplitude Oscillatory Extension”, Physical Review Fluids, 1, 053301 (2016). Web PDF
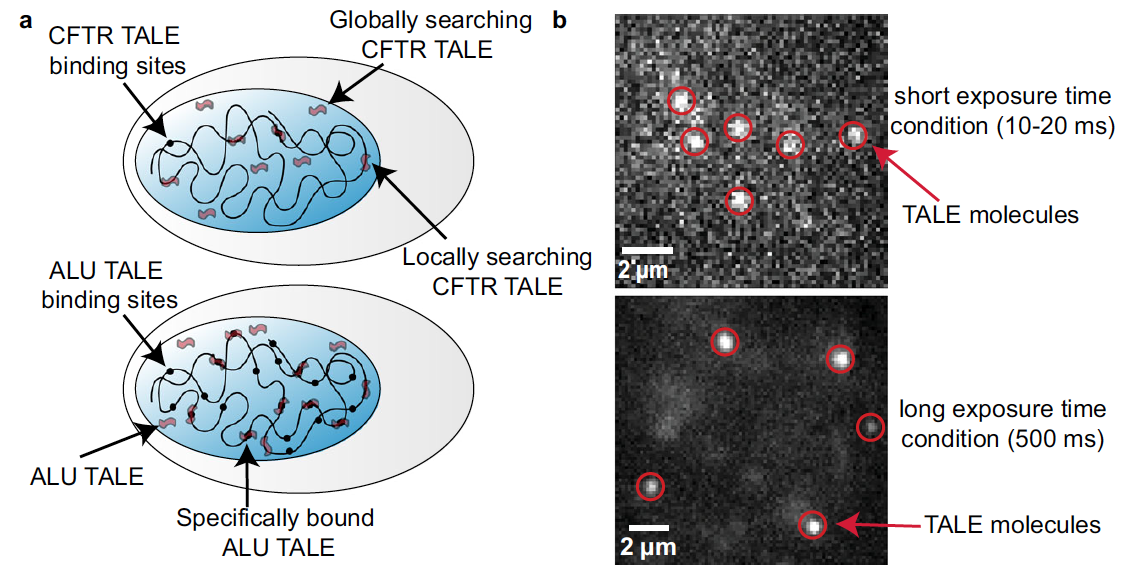
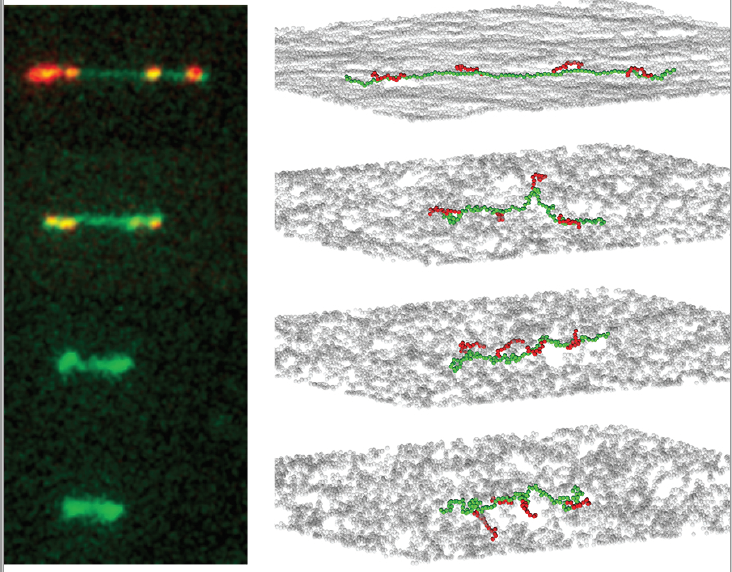
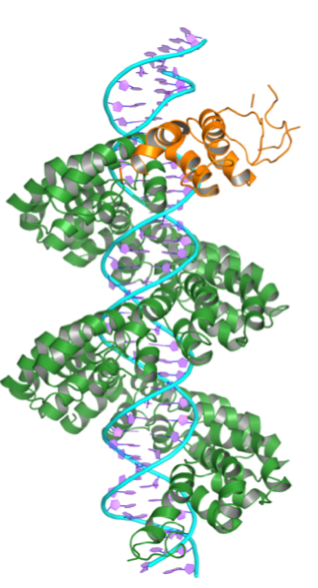
- L. W. Cuculis, Z. Abil, H. Zhao, C. M. Schroeder, “TALE Proteins Search DNA Using a Rotationally Decoupled Mechanism”, Nature Chemical Biology, 12, 831-837 (2016). Web PDF
See also: News and Views, Nature Chemical Biology
- C. M. Schroeder, S. Köster, Y. Huang, “Emerging Investigators 2016: Discovery Science Meets Technology”, Lab on a Chip, 16, 2974-2976 (2016). Web PDF
- A. Shenoy, C. V. Rao, C. M. Schroeder, “Stokes Trap for Multiplexed Particle Manipulation and Assembly Using Fluidics”, Proceedings of the National Academy of Sciences, 113, 3976-3981 (2016). Web PDF
See also:
Research Highlights Nature Physics

- K. Hsiao, C. M. Schroeder, C. E. Sing, “Ring Polymer Dynamics are Governed by a Coupling Between Architecture and Hydrodynamic Interactions”, Macromolecules, 49(5), 1961-1971, (2016). Web PDF

2015
- D. T. Reilly, S. H. Kim, J. A. Katzenellenbogen, C. M. Schroeder, “Fluorescent Nanoconjugate Derivatives with Enhanced Photostability for Single Molecule Imaging”, Analytical Chemistry, 87(21), 11048-11057, (2015). Web PDF
- Y. Li, K. Hsiao, C. A. Brockman, D.Y. Yates, R. M. Robertson-Anderson, J. A. Kornfield, M. J. San Francisco, C. M. Schroeder, G. B. McKenna, “When Ends Meet: Circular DNA Stretches Differently in Elongational Flows”, Macromolecules, 48(16), 5997–6001, (2015). Web PDF

- X. Li, C. M. Schroeder, K. D. Dorfman, “Modeling the Stretching of Wormlike Chains in the Presence of Excluded Volume”, Soft Matter, 11, 5947-5954 (2015). Web PDF

- L. W. Cuculis, Z. Abil, H. Zhao, C. M. Schroeder, “Direct Observation of TALE Protein Dynamics Reveals a Two-state Search Mechanism”, Nature Communications, 6, 7277 (2015). Web PDF

- R. Mohan, C. Sanpitakseree, A. V. Desai, S. E. Sevgen, C. M. Schroeder, P. J. A. Kenis, “A Microfluidic Approach to Study the Effect of Bacterial Interactions on Antimicrobial Susceptibility in Polymicrobial Cultures”, RSC Advances, 5, 35211-35223 (2015). Web PDF

- D. J. Mai, A. B. Marciel, C.E. Sing, C. M. Schroeder, “Topology-Controlled Relaxation Dynamics of Single Comb Polymers”, ACS Macro Letters, 4, 446-452 (2015).Web PDF

- A. B. Marciel, D. J. Mai, C. M. Schroeder, “Template-Directed Synthesis of Structurally Defined Branched Polymer Architectures”, Macromolecules, 48(5), 1296-1303 (2015). Web PDF

- A. Mukherjee and C. M. Schroeder, “Flavin-based Fluorescent Proteins: Emerging Paradigms in Biological Imaging”, Current Opinion in Biotechnology, 31, 16-23 (2015). PDF
2014
- F. Latinwo, K. Hsiao, C. M. Schroeder, “Nonequilibrium Thermodynamics of Dilute Polymer Solutions in Flow”, The Journal of Chemical Physics, 141, 174903 (2014).Web PDF

- A. Shenoy, M. Tanyeri, C. M. Schroeder, “Characterizing the Performance of the Hydrodynamic Trap Using a Control-based Approach”, Microfluidics and Nanofluidics, 8, 1055–1066(2015). Web PDF

- A. Mukherjee, K. B. Weyant, J. Walker, U. Agrawal, I. Cann, C. M. Schroeder, “Engineering and Characterization of New LOV-based Fluorescent Proteins from Chlamydomonas reinhardtii andVaucheria frigida“, ACS Synthetic Biology, DOI: 10.1021/sb500237x (2014). Web PDF

- E. M. Johnson-Chavarria, U. Agrawal, M. Tanyeri, T. E. Kuhlman, C. M. Schroeder, “Automated Single Cell Microbioreactor for Monitoring Intracellular Dynamics and Cell Growth in Free Solution”, Lab on a Chip, 14, 2688-2697 (2014). Web PDF

- K. S. Lee, A. B. Marciel, A. G. Kozlov, C. M. Schroeder, T. M. Lohman, T. Ha, “Ultrafast Re-distribution ofE. coli SSB Along Long Single-Stranded DNA via Intersegment Transfer”, Journal of Molecular Biology, 426, 2413-2421 (2014). Web PDF

- F. Latinwo and C. M. Schroeder, “Determining Elasticity from Single Polymer Dynamics”, Soft Matter, 10, 2178-2187 (2014). Web PDF

2013
- A. B. Marciel, M. Tanyeri, B. D. Wall, J. D. Tovar, C. M. Schroeder*, W. L. Wilson*, “Fluidic-directed Assembly of Aligned Oligopeptides with π -conjugated Cores”, Advanced Materials, 25, 6398-6404 (2013). Web PDF

- F. Latinwo and C. M. Schroeder, “Nonequilibrium Work Relations for Polymer Dynamics in Dilute Solutions”, Macromolecules, 46, 8345-8355 (2013). Web PDF

- M. Tanyeri and C. M. Schroeder, “Manipulation and Confinement of Single Particles using a Fluid Flow”, Nano Letters, 13, 2357-2364 (2013). Web PDF

- R. Mohan*, A. Mukherjee*, S. E. Sevgen, C. Sanpitakseree, J. Lee, C. M. Schroeder, P. J. A. Kenis, “A Multiplexed Microfluidic Platform for Rapid Antibiotic Susceptibility Testing”, Biosensors and Bioelectronics, 49, 118-125 (2013). Web PDF

- A. Mukherjee, J. Walker, K. B. Weyant, C. M. Schroeder, “Characterization of Flavin-based Fluorescent Proteins: An Emerging Class of Powerful Fluorescent Reporters”, PLOS ONE, 8, e64753 (2013). Web PDF

- A. B. Marciel and C. M. Schroeder, “New Directions in Single Polymer Dynamics”, Journal of Polymer Science: Polymer Physics, 51, 556-566 (2013). Web PDF

- U. Agrawal, D. Reilly, C. M. Schroeder, “Zooming in on Biological Processes with Fluorescence Nanoscopy”, Current Opinion in Biotechnology, 24 (2013). Web PDF

- Y. Kim, S. Kim, M. Tanyeri, J. A. Katzenellenbogen, C. M. Schroeder, “Dendrimer Probes for Enhanced Photostability and Localization in Fluorescence Imaging”, Biophysical Journal, 104, 1566-1575, (2013).
Web PDF
News: This article was highlighted in a “New and Notable” article: Biophysical Journal, 104, 1394 (2013).

2012
- A. Mukherjee and C. M. Schroeder, “Directed Evolution of Bright Mutants of a Flavin-Dependent Anaerobic Fluorescent Protein from Pseudomonas putida“, Journal of Biological Engineering, 6, 20, (2012). Web PDF

- Y. Kim, S. Kim, J. A. Katzenellenbogen, C. M. Schroeder, “Specific Labeling of Zinc Finger Proteins using Non-canonical Amino Acids and Copper-free Click Chemistry”, Bioconjugate Chemistry, 23, 1891-1901 (2012). Web PDF

- D. J. Mai, C. A. Brockman, C. M. Schroeder, “Microfluidic Systems for Single DNA Dynamics”, Soft Matter, 8,10560-10572 (2012). Web PDF

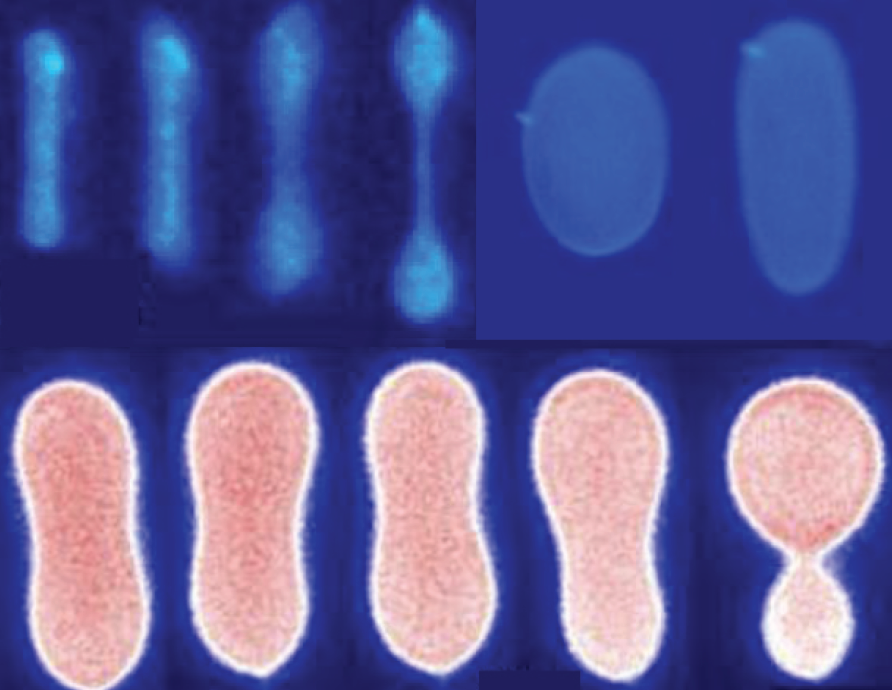
- M.-H. Lai, J. H. Jeong, R. Devolder, C. Brockman, C. M. Schroeder, H. Kong, “Ellipsoidal Polyaspartamide Polymersomes with Enhanced Cell-Targeting Ability”, Advanced Functional Materials, 22, 3239-3246 (2012). Web PDF

2011
- M. Tanyeri, M. Ranka, N. Sittipolkul, C. M. Schroeder, “Microfluidic Wheatstone Bridge for Rapid Sample Analysis”, Lab on a Chip, 11, 4181-4186 (2011). Web PDF

- F. Latinwo and C. M. Schroeder, “Model Systems for Single Molecule Polymer Dynamics”, Soft Matter, 7, 7907-7913 (2011), June (2011). Web PDF
News: This article was selected as a “Hot Article” by the editor of Soft Matter.

- C. A. Brockman, S. Kim, F. Latinwo, C. M. Schroeder, “Direct Observation of Single Flexible Polymers using Single Stranded DNA”, Soft Matter, 7, 8005-8012 (2011). Web PDF
News: This article was selected as a “Hot Article” by the editor of Soft Matter.

- M. Tanyeri, M. Ranka, N. Sittipolkul, C. M. Schroeder, “A Microfluidic-based Hydrodynamic Trap: Design and Implementation”, Lab on a Chip, 11, 1786-1794 (2011). Web PDF

- B. Schudel, M. Tanyeri, A. Mukherjee, C. M. Schroeder, P. J. A. Kenis, “Multiplexed Detection of Nucleic Acids in a Combinatorial Screening Chip”, Lab on a Chip, 11, 1916-1923 (2011). Web PDF

- E. M. Johnson-Chavarria , M. Tanyeri , C. M. Schroeder, “A Microfluidic-based Hydrodynamic Trap for Single Particles”, Journal of Visualized Experiments, 47, (2011). Web PDF
2010
- Y. Han, D. Dodd, C. M. Schroeder, R. I. Mackie, I. K. O. Cann, “Comparative Analysis of Two Thermophilic Enzymes Exhibiting both β-1,4-Mannosidic and β-1,4-Glucosidic Cleavage Activities fromCaldanaerobius polysaccharolyticus“, Journal of Bacteriology, 192, 4111-4121 (2010). Web PDF

- M. Tanyeri, E. M. Johnson-Chavarria and C. M. Schroeder, “Hydrodynamic Trap for Single Particles and Cells”, Applied Physics Letters, 96, 224101 (2010). Web PDF

- S. Kim, C. M. Schroeder, X. S. Xie, “Single-Molecule Study of DNA Polymerization Activity of HIV-1 Reverse Transcriptase on DNA Templates”, Journal of Molecular Biology, 395, 995-1006 (2010). Web PDF

- C. J. Yeoman, Y. Han, D. Dodd, C. M. Schroeder, R. I. Mackie, I. K. O. Cann, “Thermostable Enzymes as Biocatalysts in the Biofuel Industry”, Advances in Applied Microbiology, 70 (2010). Web PDF

2009 (Prior to Illinois)
- S. Kim, P. C. Blainey, C. M. Schroeder, X. S. Xie, “Multiplexed Single-molecule Assay for Enzymatic Activity on Flow-stretched DNA”, Nature Methods, 4, 397-399 (2007). Web PDF
- C. M. Schroeder, R. E. Teixeira, E. S. G. Shaqfeh, S. Chu, “Characteristic Periodic Motion of Polymers in Shear Flow”, Physical Review Letters, 95, 018301 (2005). Web PDF

- C. M. Schroeder, R. E. Teixeira, E. S. G. Shaqfeh, S. Chu, “Dynamics of DNA in the Flow-Gradient Plane of Steady Shear Flow: Observations and Simulations”, Macromolecules, 38, 1967-1978 (2005).
Web PDF

- C. M. Schroeder, E. S. G. Shaqfeh, S. Chu, “Effect of Hydrodynamic Interactions on DNA Dynamics in Extensional Flow: Simulation and Single Molecule Experiment”, Macromolecules, 37, 9242-9256(2004). Web PDF

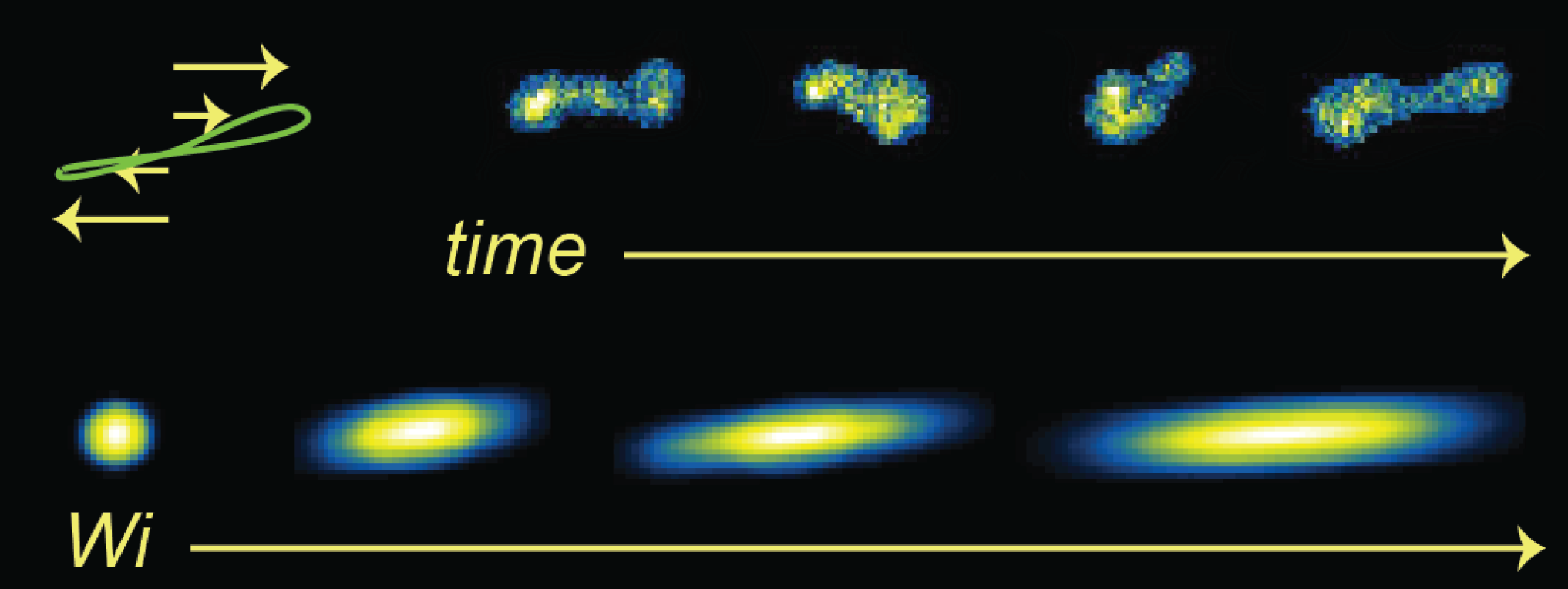
- C. M. Schroeder, H. P. Babcock, E. S. G. Shaqfeh, S. Chu, “Observation of Polymer Conformation Hysteresis in Extensional Flow”, Science, 301, 1515-1519 (2003). Web PDF

- S. J. Vinay, D. M. Phillips, Y. S. Lee, C. M. Schroeder, X. Ma, M. C. Kim, M. S. Jhon, “Simulation of Ultrathin Lubricant Films Spreading of Various Carbon Surfaces”, Journal of Applied Physics, 87, 6164-6166 (2000). Web PDF
Book Chapters Back to Top
- C. M. Schroeder, P. C. Blainey, S. Kim, X. S. Xie, “Hydrodynamic Flow-stretching Assay for Single Molecule Studies of Nucleic Acid-Protein Interactions”, in Single Molecule Techniques: A Laboratory Manual, T. Ha and P. Selvin (eds.), Cold Spring Harbor Laboratory Press, 2007.
- A. Mukherjee and C. M. Schroeder, “Microfluidic Methods in Single Cell Biology”, in Microfluidic Methods in Molecular Biology , C. Lu and S. Verbridge (eds.), Springer, in press, 2016.
Patents Back to Top
- C. M. Schroeder, H. P. Babcock, E. S. G. Shaqfeh, S. Chu, “System and Method for Confining an Object to a Region of Fluid Flow Having a Stagnation Point”, United States Patent, No. 7,013,739 B2, March 21, 2006.
- Y. Kim, S. Kim, M. Tanyeri, J. A. Katzenellenbogen, C. M. Schroeder, “Dye-conjugated Dendrimers”, United States Patent, No. 13/385,828, University of Illinois at Urbana-Champaign, May 2016.