The cutting edge of chemical science research lies in the ability to manipulate and control single molecules. The Schroeder Group uses single-molecule characterization of materials — together with automated synthesis and closed-loop materials discovery — to understand how form and function arise in soft materials. This powerful approach allows for the precise design and development of new functional materials, in problems ranging from lipid membrane dynamics to bioelectronic materials.
Research in the Schroeder Group is focused on four areas: (1) automated synthesis for closed-loop materials discovery, including “Lego-like” or modular synthesis of organic materials using molecular building blocks, (2) molecular electronics of conjugated molecules, light-harvesting organic molecules, and redox-active materials, in addition to protein nanowires and bioelectronic materials, (3) dynamics of lipid membranes, vesicles, and colloidal particles, including new methods to deform and study soft materials using the Stokes trap, and (4) single polymer dynamics, including entangled solutions and architecturally complex polymers such as rings and branched polymers.
Research Highlights
Automated synthesis & materials discovery (more…)
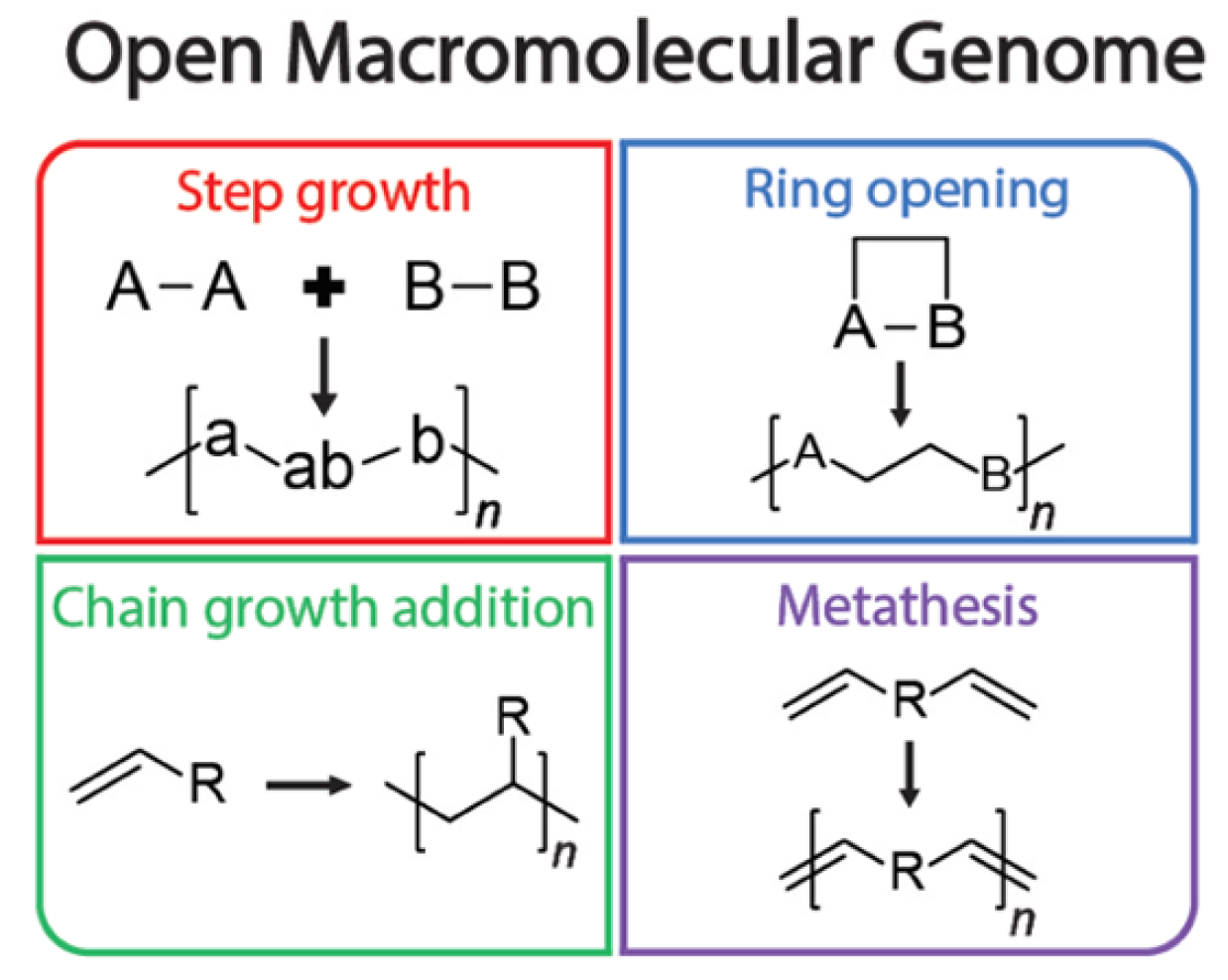
Open macromolecular genome: generative design of synthetically accessible polymers |
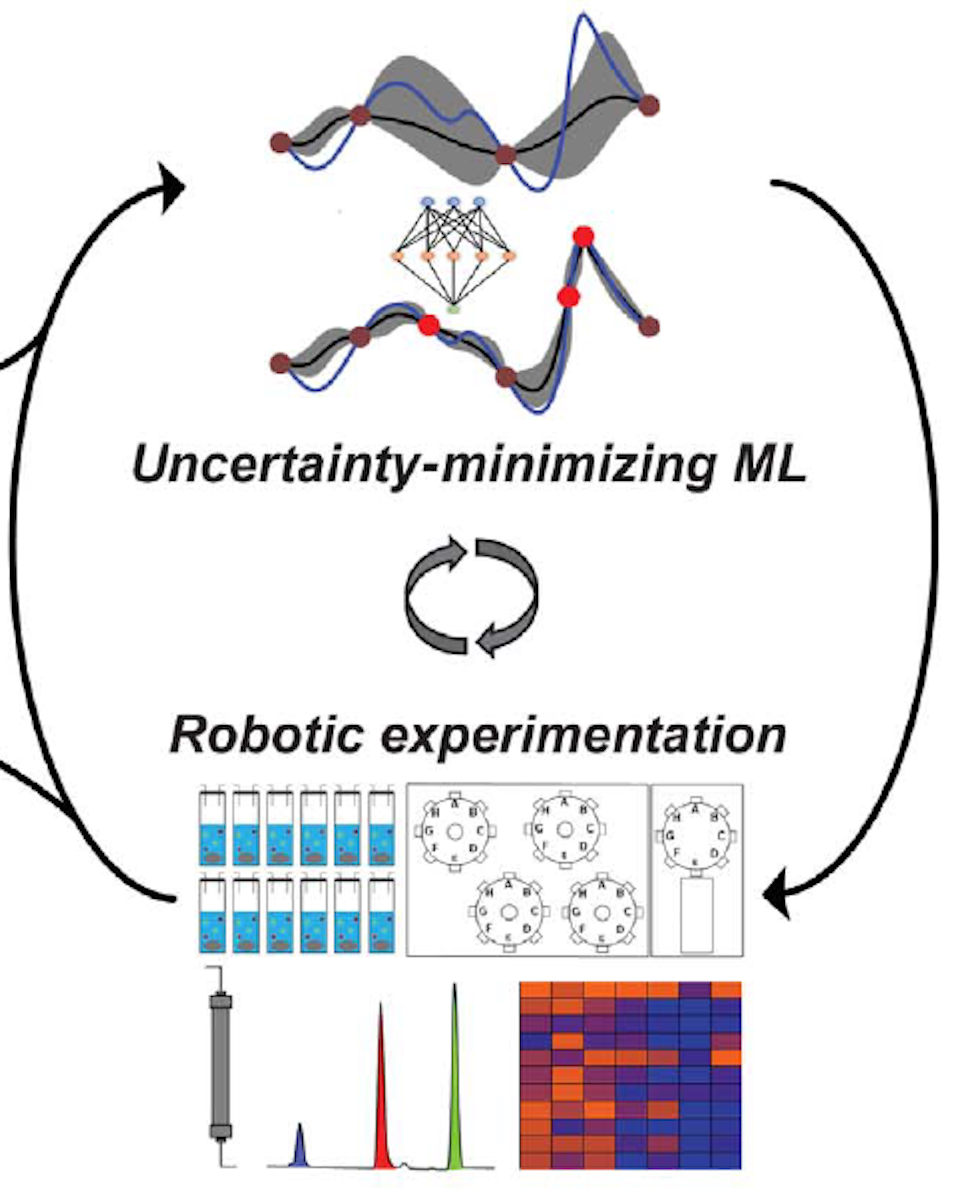
Closed-loop optimization of general reaction conditions |
Automated synthesis for single-molecule electronics |
||
 |
 |
 |
||
| Science, 2022 | Nature Communications, 2022 |
Molecular electronics (more…)
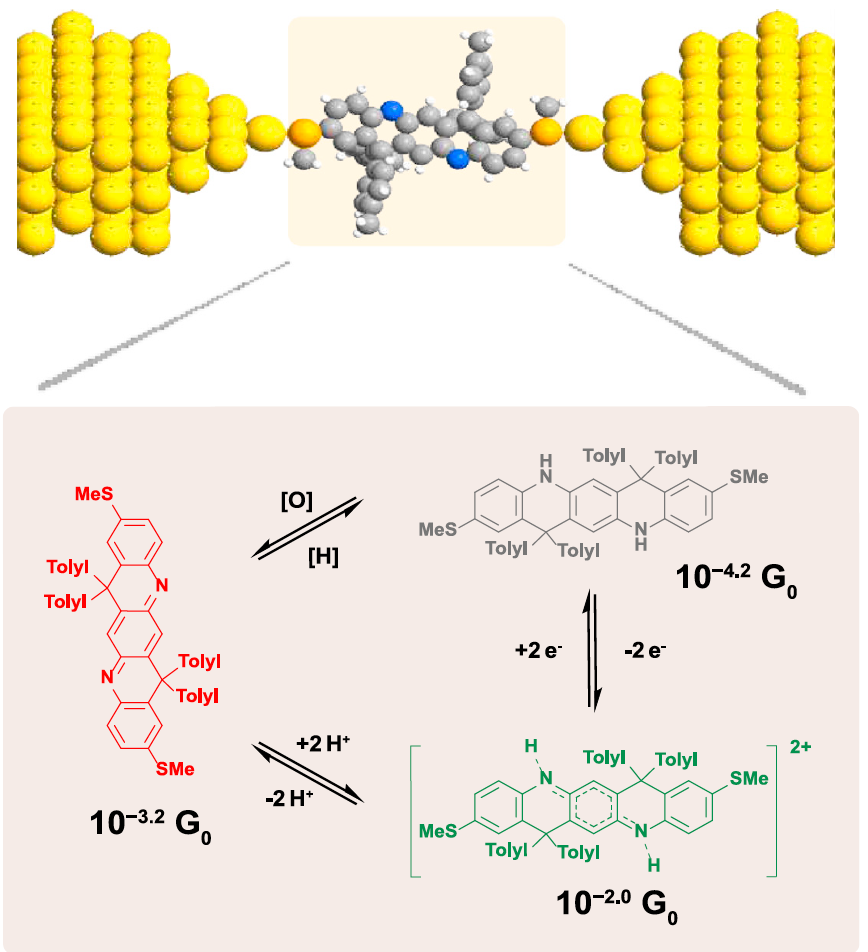
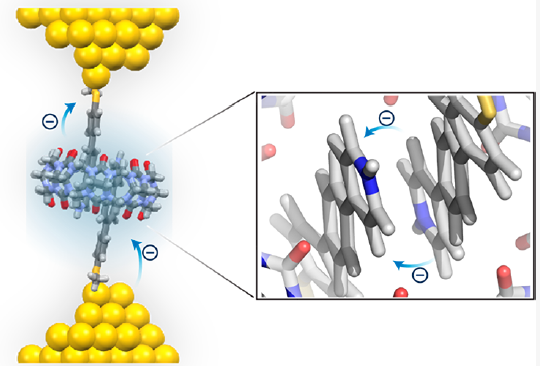
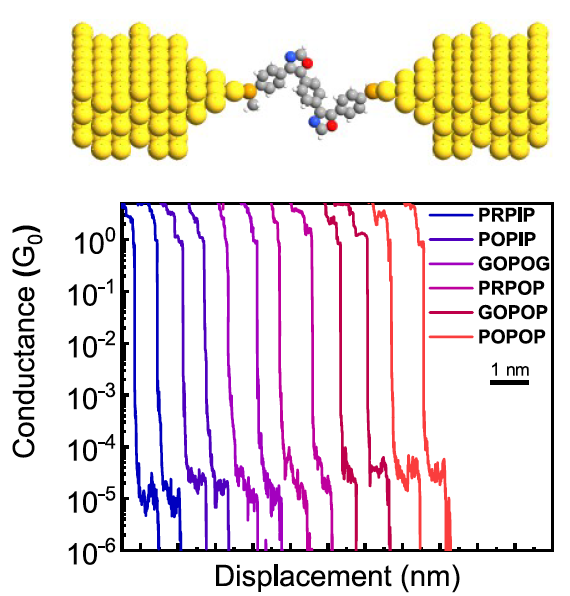
Ladder-type molecules as single-molecule electronic switches |
Single-molecule electronics with host-guest complexes |
Charge transport in sequence-defined oligomers |
||
 |
 |
 |
||
| JACS, 2022 | JACS, 2020 |
Vesicle & lipid membrane dynamics (more…)
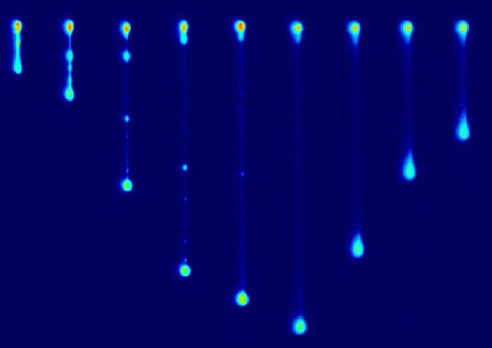
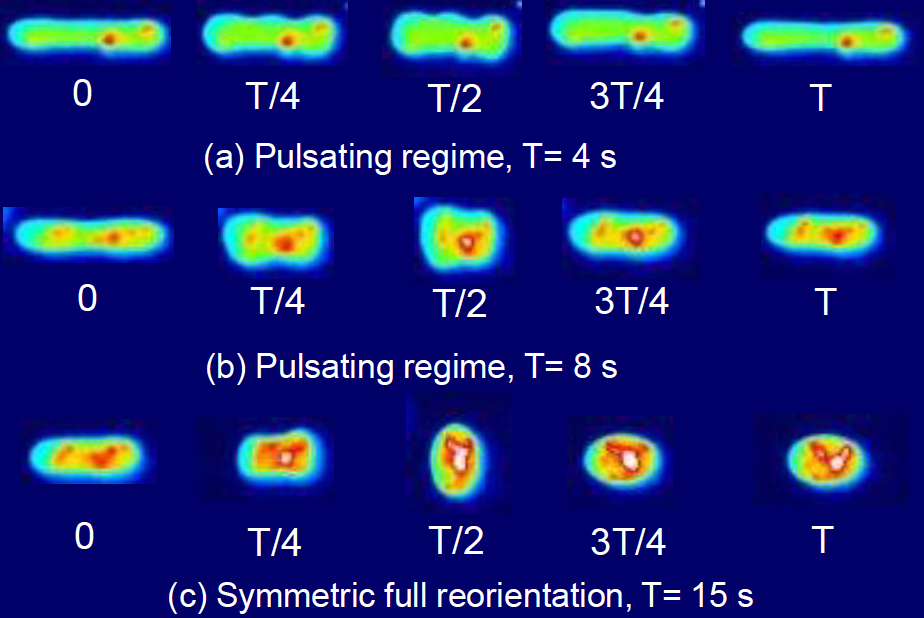
Non-equilibrium stretching of vesicles in flow |
Dynamics of vesicles in oscillatory flows |
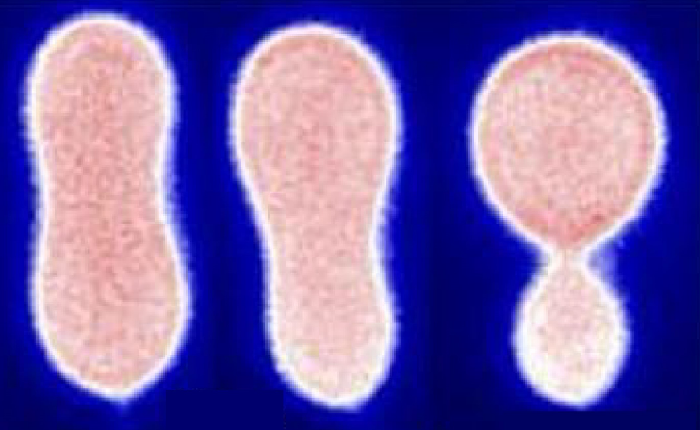
Flow-phase diagram for lipid vesicles in extensional flow |
||
 |
 |
 |
||
| Journal of Fluid Mechanics, 2021 | Soft Matter, 2020 |
Stokes Trap (more…)
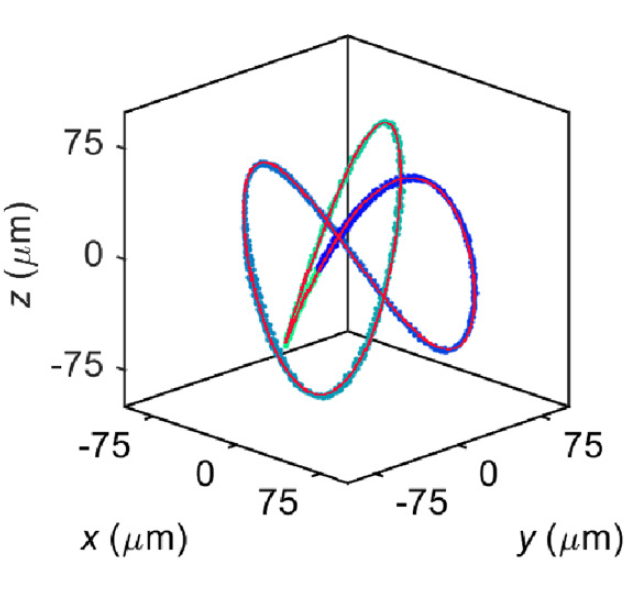
3D manipulation and dynamics of soft materials in 3D flows |
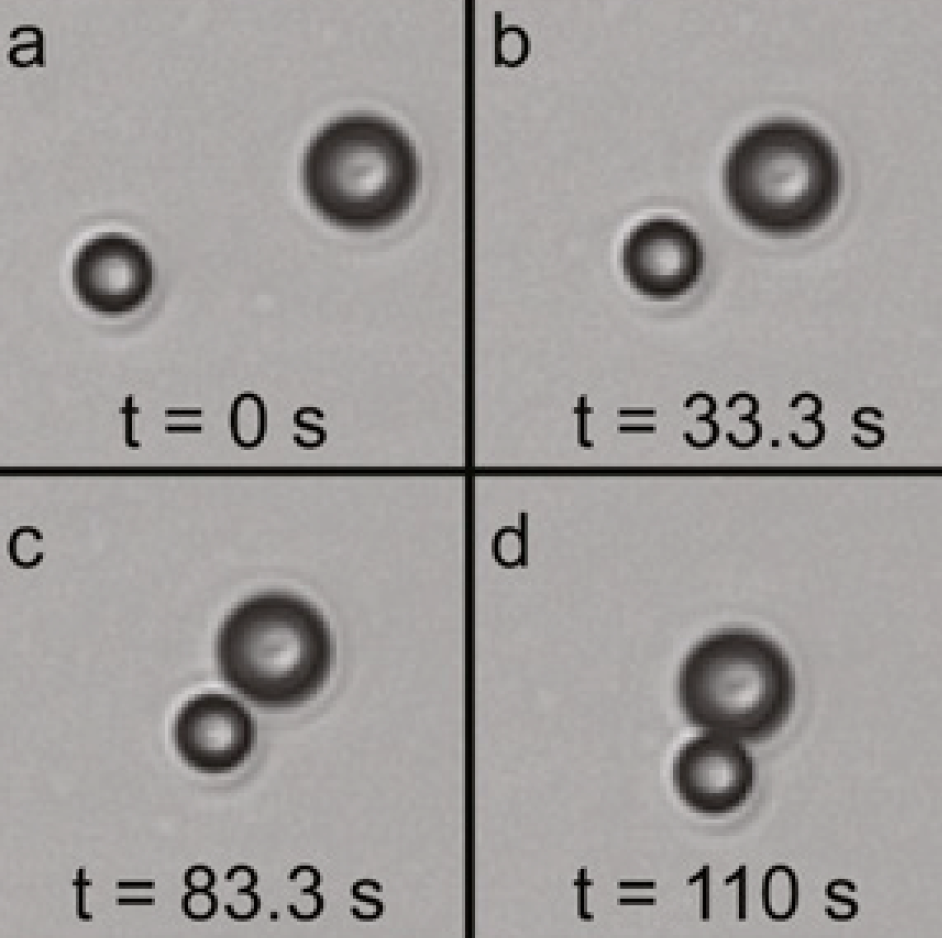
Orientation control and trajectory tracking with automated flow control |
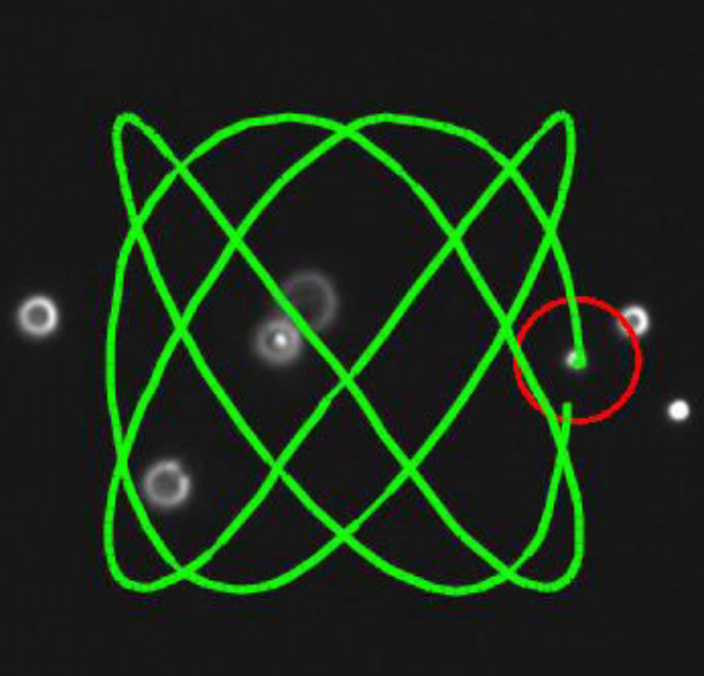
Stokes trap for multiplexed particle manipulation |
||
 |
 |
 |
||
| Physical Review Fluids, 2019 | PNAS, 2016 |
Single polymer dynamics (more…)
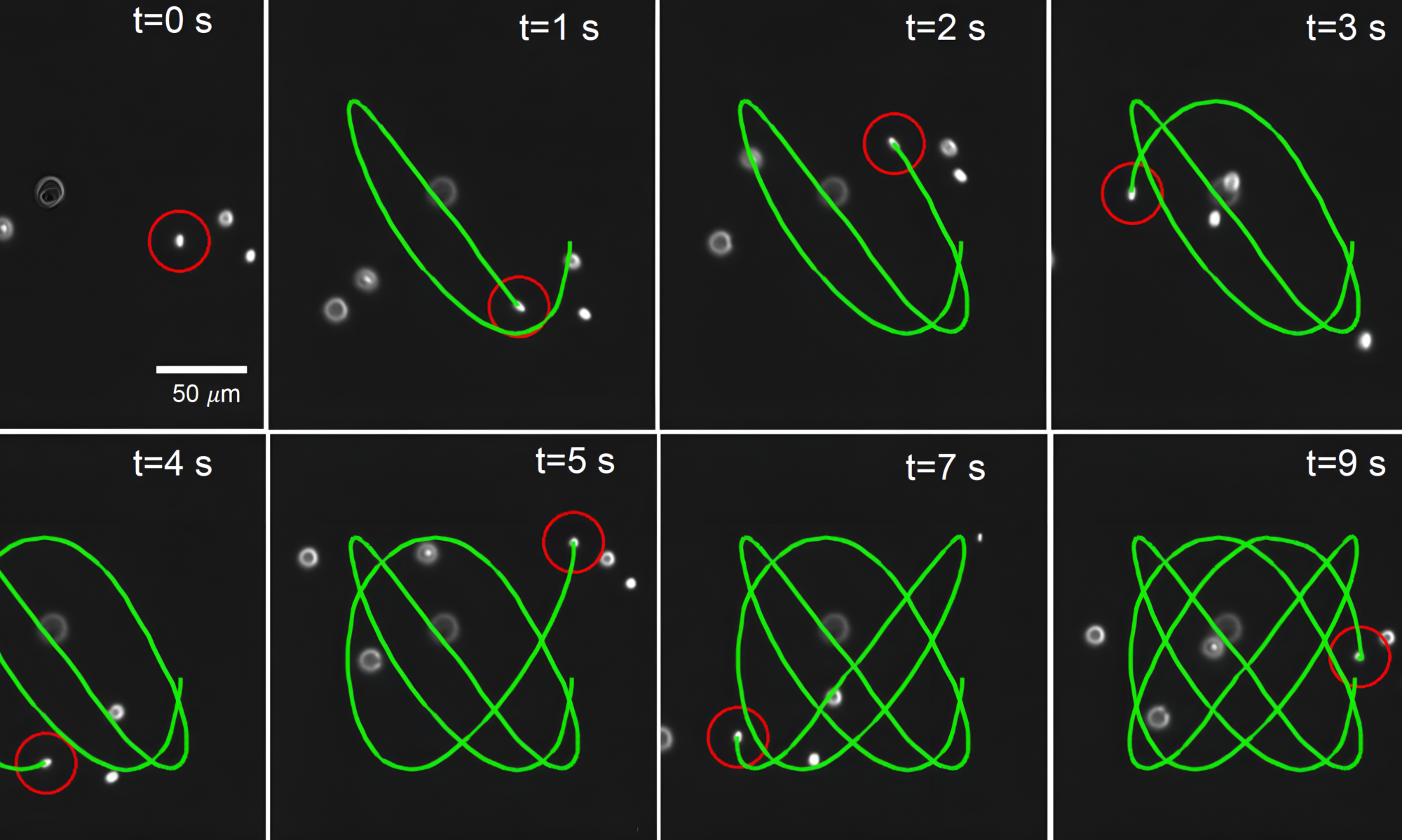
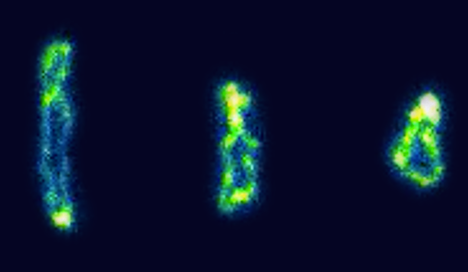
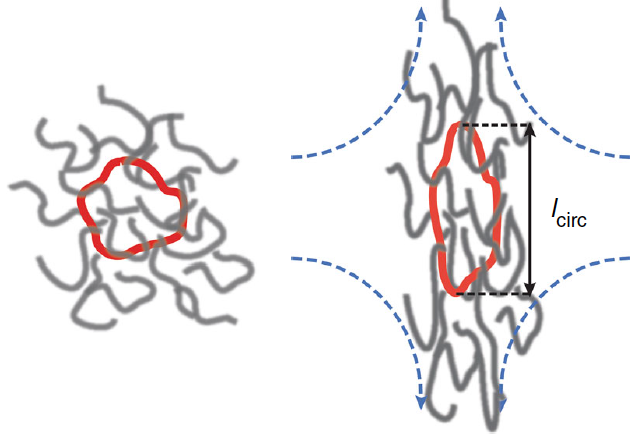
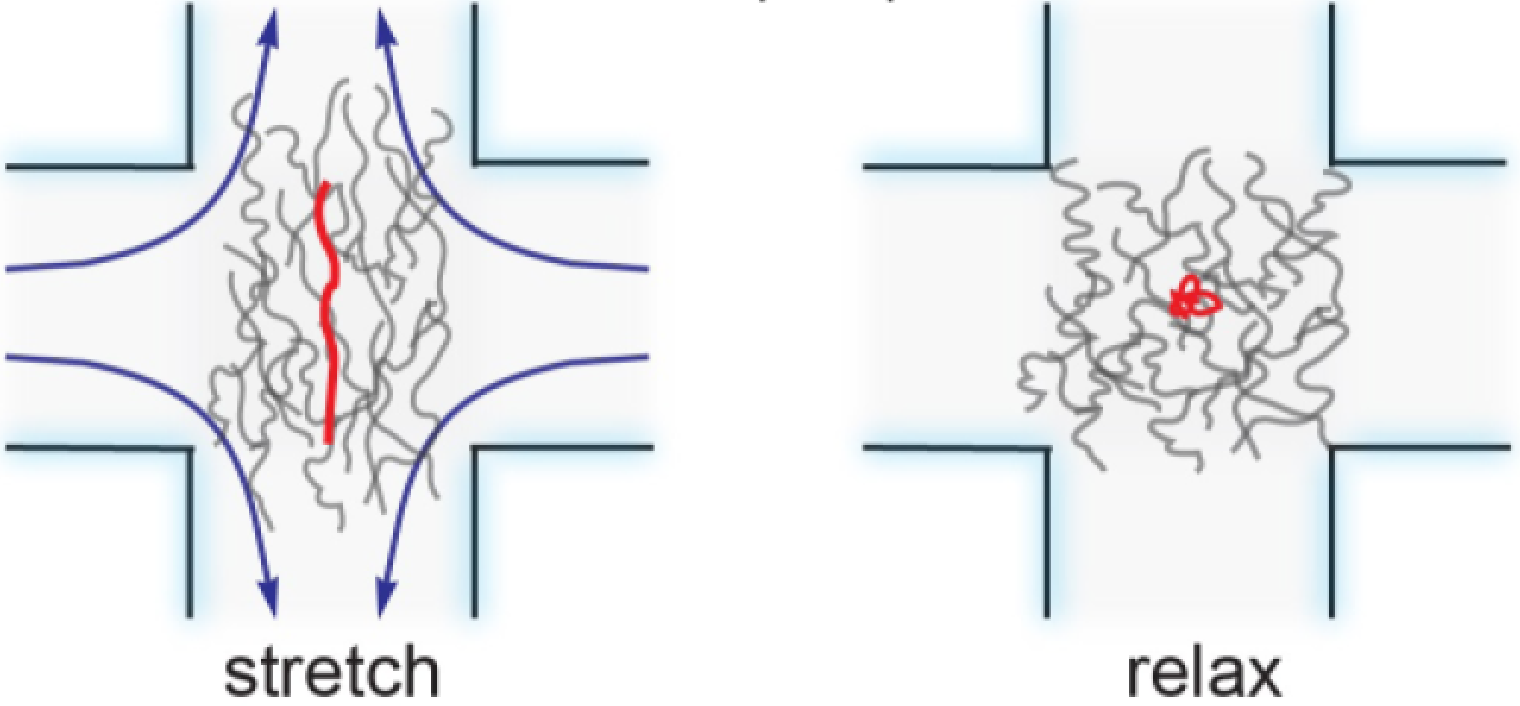
Direct observation of ring polymer dynamics in shear flow |
Ring-linear threading events in semi-dilute solutions |
Dynamically heterogeneous relaxation of entangled polymer solutions |
||
 |
 |
 |
||
| Nature Communications, 2019 | PRL, 2018 |
Prior Research
Supramolecular assembly of proteins
Single molecule biophysics & fluorescent proteins