Overview: We use single molecule techniques to directly observe the dynamics of single polymer chains far from equilibrium. Our work establishes important connections between molecular-scale phenomena and macroscopic properties in soft materials. In recent years, our group has advanced the field of single polymer dynamics to understand the molecular rheology and dynamics of polymers with complex architectures (stars, combs, rings, bottlebrushes), different backbone chemistries (single stranded DNA, PNIPAM-DNA copolymers, chemically modified DNA), and entangled and semi-dilute polymer solutions.
Selected publication:
C. M. Schroeder, “Single Polymer Dynamics for Molecular Rheology”, Journal of Rheology, 62, 371 (2018).
Single molecule studies of ring polymers
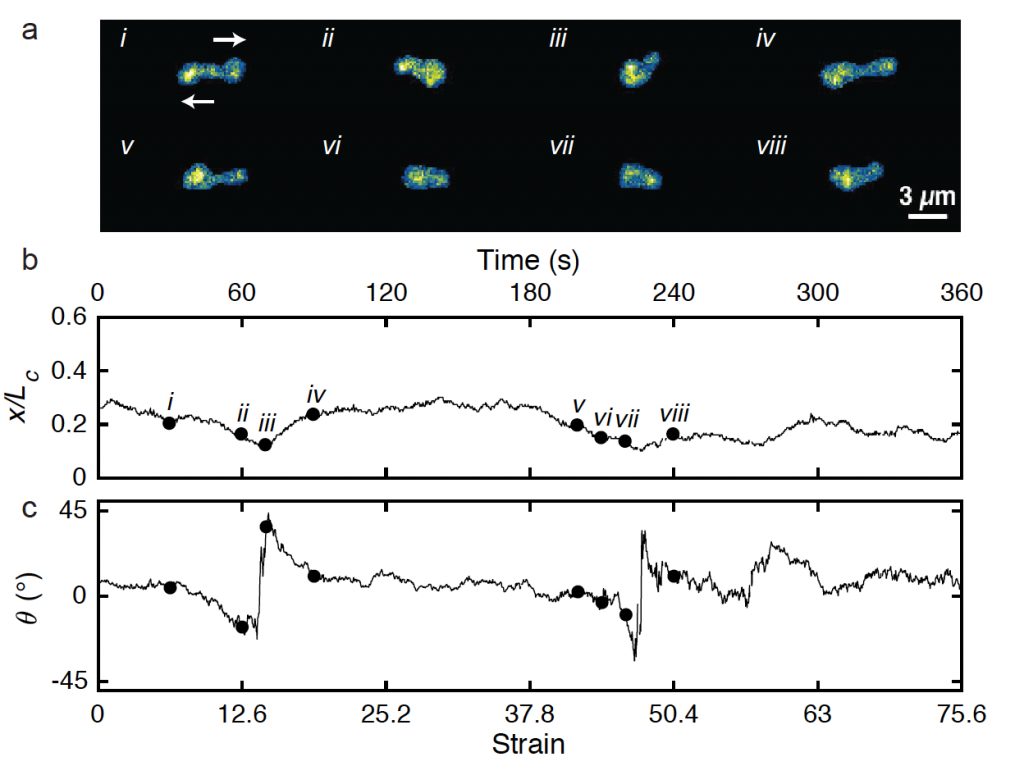 Ring polymers are a unique class of macromolecules that lack free ends, which gives rise to qualitatively different flow behavior compared to linear polymers. Circular macromolecules play a key role in biology, including replication of bacterial genomes, plasmid-based DNA vaccines, and biologically active macrocycles as drugs. Circular macromolecules call into question our basic understanding of polymer dynamics because the absence of free ends alters chain dynamics, diffusion, and ordering transitions compared to linear macromolecules. Our work has provided some of the first direct observations of ring polymer dynamics in flow, revealing unexpected behavior due to the closed ring architecture. We observed the conformational dynamics of ring DNA molecules in extensional flow using single molecule techniques. Our results show that ring polymers exhibit a shifted coil-to-stretch transition and less diverse molecular individualism compared to linear chains. We further studied the dynamics of single rings in semi-dilute solutions of linear polymers and ring-linear blends. In addition, we directly visualized the dynamics of single ring polymers in shear flow using custom-built flow devices, thereby revealing ring stretching and tumbling dynamics in shear flow. Our work has provided fundamentally new information on the non-equilibrium dynamics of ring polymers in flow.
Ring polymers are a unique class of macromolecules that lack free ends, which gives rise to qualitatively different flow behavior compared to linear polymers. Circular macromolecules play a key role in biology, including replication of bacterial genomes, plasmid-based DNA vaccines, and biologically active macrocycles as drugs. Circular macromolecules call into question our basic understanding of polymer dynamics because the absence of free ends alters chain dynamics, diffusion, and ordering transitions compared to linear macromolecules. Our work has provided some of the first direct observations of ring polymer dynamics in flow, revealing unexpected behavior due to the closed ring architecture. We observed the conformational dynamics of ring DNA molecules in extensional flow using single molecule techniques. Our results show that ring polymers exhibit a shifted coil-to-stretch transition and less diverse molecular individualism compared to linear chains. We further studied the dynamics of single rings in semi-dilute solutions of linear polymers and ring-linear blends. In addition, we directly visualized the dynamics of single ring polymers in shear flow using custom-built flow devices, thereby revealing ring stretching and tumbling dynamics in shear flow. Our work has provided fundamentally new information on the non-equilibrium dynamics of ring polymers in flow.
Selected publications:
- Y. Zhou, C. D. Young, K. E. Regan, M. Lee, S. Banik, D. Kong, G. B. McKenna, R. M. Robertson-Anderson, C. E. Sing, C. M. Schroeder, “Dynamics and Rheology of Ring-Linear Blend Semidilute Solutions in Extensional Flow: Single Molecule Experiments”, Journal of Rheology, 65, 729-744 (2021).
- C. D. Young, Y. Zhou, C. M. Schroeder, C. E. Sing, “Dynamics and Rheology of Ring-Linear Blend Semidilute Solutions in Extensional Flow: Modeling and Molecular Simulations”, Journal of Rheology, 65, 757-777 (2021).
- M. Tu, M. Lee, R. M. Robertson-Anderson, C. M. Schroeder, “Direct Observation of Ring Polymer Dynamics in the Flow-Gradient Plane of Shear Flow”, Macromolecules, 53, 9406–9419 (2020).
- Y. Zhou, K. W. Hsiao, K. E. Regan, D. Kong, G. B. McKenna, R. M. Robertson-Anderson, C. M. Schroeder, “Dynamics of Single Ring Polymers in Semi-dilute Linear Polymer Solutions”, Nature Communications, 10, 1753 (2019).
- K. Hsiao, C. M. Schroeder, C. E. Sing, “Ring Polymer Dynamics Are Governed by a Coupling between Architecture and Hydrodynamic Interactions”, Macromolecules, 49, 1961 (2016).
- Y. Li, K. Hsiao, C.A. Brockman, D.Y. Yates, R.M. Robertson-Anderson, J.A. Kornfield, M.J. San Francisco, C. M. Schroeder, G. B. McKenna, “When Ends Meet: Circular DNA Stretches Differently in Elongational Flows”, Macromolecules, 48, 5997 (2015).
Single molecule studies of branched polymers
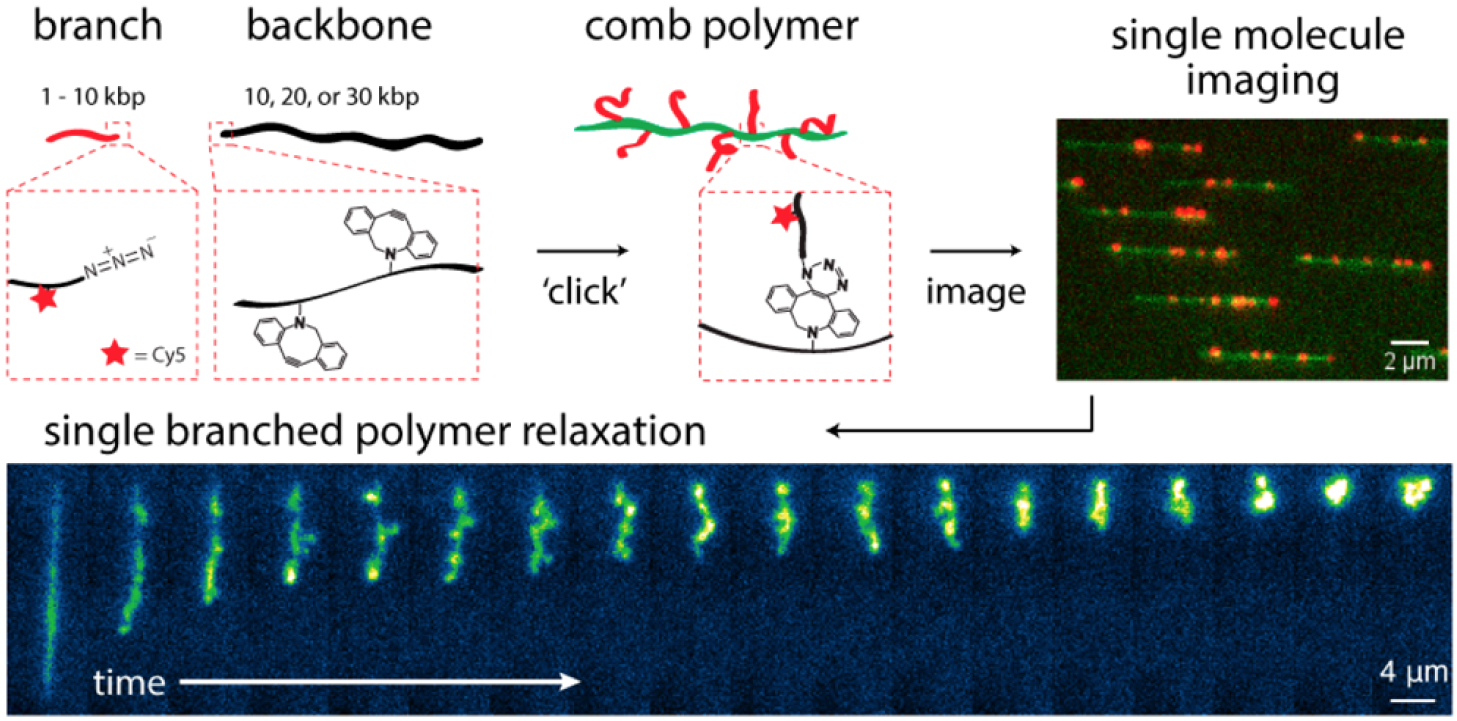 In one project, we synthesized and directly observed the dynamics of DNA comb polymers using single molecule techniques. Our results show that the molecular architecture of branched polymers plays unexpected roles in the relaxation and stretching dynamics of polymers in flow. DNA comb polymers are synthesized using an enzymatic-synthetic approach, wherein chemically modified DNA branches and DNA backbones are generated in separate polymerase chain reactions, followed by a ‘graft-onto’ reaction via strain-promoted [3+2] azide-alkyne cycloaddition. Single molecule fluorescence microscopy is then used to directly visualize branched polymers, such that the backbone and side branches can be tracked independently using dual-color fluorescence labeling. Our work has pioneered the direct observation of comb polymer relaxation from high stretch, stretching dynamics in extensional flow, and dynamics in semi-dilute polymer solutions.
In one project, we synthesized and directly observed the dynamics of DNA comb polymers using single molecule techniques. Our results show that the molecular architecture of branched polymers plays unexpected roles in the relaxation and stretching dynamics of polymers in flow. DNA comb polymers are synthesized using an enzymatic-synthetic approach, wherein chemically modified DNA branches and DNA backbones are generated in separate polymerase chain reactions, followed by a ‘graft-onto’ reaction via strain-promoted [3+2] azide-alkyne cycloaddition. Single molecule fluorescence microscopy is then used to directly visualize branched polymers, such that the backbone and side branches can be tracked independently using dual-color fluorescence labeling. Our work has pioneered the direct observation of comb polymer relaxation from high stretch, stretching dynamics in extensional flow, and dynamics in semi-dilute polymer solutions.
Selected publications:
- S. Patel, C. D. Young, C. E. Sing, C. M. Schroeder, “Non-monotonic Dependence of Comb Polymer Relaxation on Branch Density in Semi-dilute Solutions”, Physical Review Fluids, 5, 121301R (2020).
- D. J. Mai, A. Sadaat, B. Khomami, C. M. Schroeder, “Stretching Dynamics of Single Comb Polymers in Extensional Flow, Macromolecules, 51, 1507 (2018).
- S. Li and C. M. Schroeder, “Synthesis and Single Molecule Studies of Thermo-responsive DNA Copolymers”, ACS Macro Letters, 7, 281 (2018).
- D. Mai, A. B. Marciel, C. E. Sing, C. M. Schroeder, “Topology-Controlled Relaxation Dynamics of Single Branched Polymers”, ACS Macro Letters, 4, 446 (2015).
- A. B. Marciel, D. J. Mai, C. M. Schroeder, “Template-directed Synthesis of Structurally Defined Branched Polymers”, Macromolecules, 48, 1296 (2015).
Single polymer dynamics in entangled and semi-dilute solutions
 The dynamics of semi-dilute polymer solutions is an intriguing yet particularly challenging problem in soft materials and rheology. Semi-dilute polymer solutions exhibit large concentration fluctuations, which precludes straightforward treatment using a mean-field approach. We used single molecule techniques to study the dynamics of semi-dilute DNA solutions in extensional flow, including polymer relaxation from high stretch, transient stretching dynamics, and steady-state stretching in flow. Interestingly, we observe a unique set of molecular conformations during the transient stretching process for single polymers in semi-dilute solutions, which suggests that the transient stretching pathways for polymer chains in semi-dilute solutions is qualitatively different compared to dilute solutions due to intermolecular interactions.
The dynamics of semi-dilute polymer solutions is an intriguing yet particularly challenging problem in soft materials and rheology. Semi-dilute polymer solutions exhibit large concentration fluctuations, which precludes straightforward treatment using a mean-field approach. We used single molecule techniques to study the dynamics of semi-dilute DNA solutions in extensional flow, including polymer relaxation from high stretch, transient stretching dynamics, and steady-state stretching in flow. Interestingly, we observe a unique set of molecular conformations during the transient stretching process for single polymers in semi-dilute solutions, which suggests that the transient stretching pathways for polymer chains in semi-dilute solutions is qualitatively different compared to dilute solutions due to intermolecular interactions.
Selected publications:
- Y. Zhou and C. M. Schroeder, “Dynamically Heterogeneous Relaxation of Entangled Polymer Chains”, Physical Review Letters, 120, 267801 (2018).
- K. Peddireddy, M. Lee, Y. Zhou, S. Adalbert, C. M. Schroeder, R. Robertson-Anderson, “Unexpected Entanglement Dynamics in Semidilute Blends of Supercoiled and Ring DNA”, Soft Matter, 16, 152 (2020).
- K. Hsiao, C. Sasmal, J. R. Prakash, C. M. Schroeder, “Direct Observation of DNA Dynamics in Semi-dilute Solutions in Extensional Flow”, Journal of Rheology, 61, 151 (2017).
- C. Sasmal, K. Hsiao, C. M. Schroeder, J. R. Prakash, “Parameter-free Prediction of DNA Dynamics in Planar Extensional Flow of Semi-dilute Solutions”, Journal of Rheology, 61, 169 (2017).
Single polymer dynamics in oscillatory flows
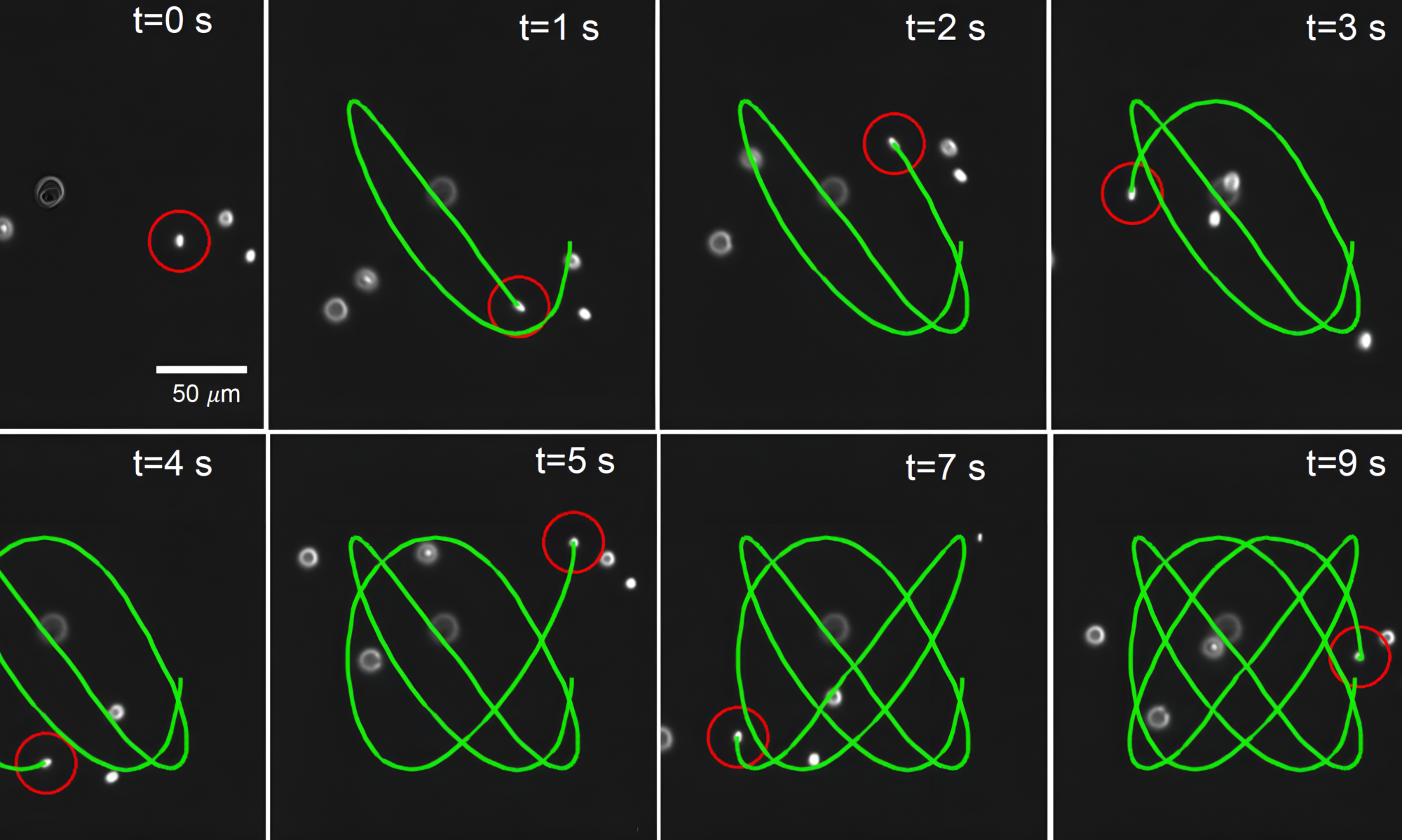
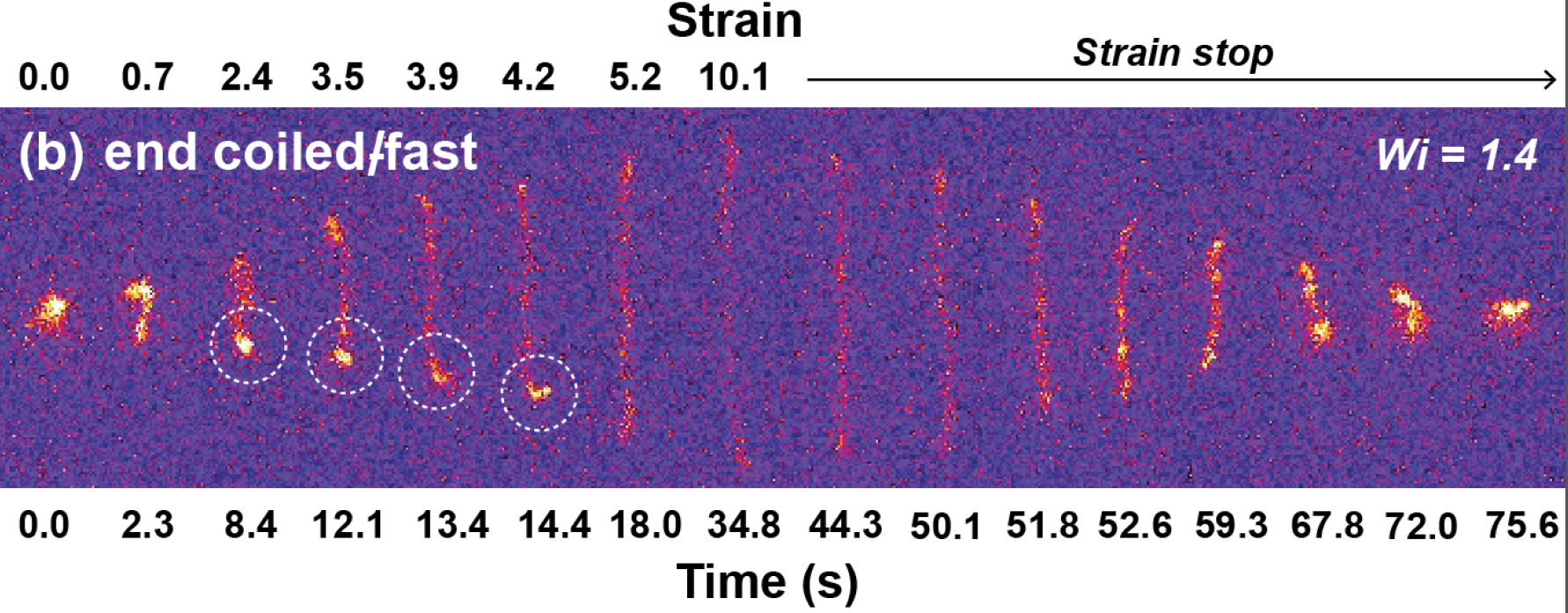
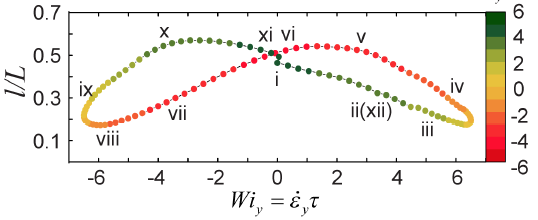 We further studied the dynamics of single polymers in large amplitude oscillatory extensional (LAOE) flow. Our results show that polymers experience periodic cycles of compression, re-orientation, and extension with unique molecular conformations in time-dependent flows. For the first time, our work enabled determination of single polymer Lissajous curves (molecular stretch-strain rate plots) to characterize both the linear and nonlinear responses as functions of dimensionless strength (Weissenberg number Wi) and probing frequency (Deborah number De).
We further studied the dynamics of single polymers in large amplitude oscillatory extensional (LAOE) flow. Our results show that polymers experience periodic cycles of compression, re-orientation, and extension with unique molecular conformations in time-dependent flows. For the first time, our work enabled determination of single polymer Lissajous curves (molecular stretch-strain rate plots) to characterize both the linear and nonlinear responses as functions of dimensionless strength (Weissenberg number Wi) and probing frequency (Deborah number De).
Selected publications:
- Y. Zhou and C. M. Schroeder, “Transient and Average Unsteady Dynamics of Single Polymers in Large-amplitude Oscillatory Extension”, Macromolecules, 49, 8018 (2016).
- Y. Zhou and C. M. Schroeder, “Single Polymer Dynamics Under Large Amplitude Oscillatory Extension”, Physical Review Fluids, 1, 053301 (2016).