Understanding the dynamics of gene editing proteins at the molecular level
 Recent advances in genome engineering offer the potential to dramatically alter the treatment of human disease. Achieving this potential, however, remains challenging due to the high degrees of precision and accuracy required for modifying large, intact genomes. Our group has used single molecule techniques to directly study the search process for transcription activator-like effector nucleases or TALE(N)s along DNA molecules, thereby revealing the molecular mechanisms underlying the DNA search process for these proteins. We use single molecule experiments to study TALE search along DNA based total internal reflection microscopy.Unexpectedly, our results show that TALEs utilize a two-state ‘search and check’ model for finding target sites while moving along DNA using a rotationally decoupled mechanism, despite remaining associated with DNA templates during the search process. In this way, TALE search is largely absent of rotationally coupled sliding. Our results suggest that the helical structure of TALEs enables these proteins to adopt a loose wrapped conformation around DNA templates during non-specific search, thereby facilitating rapid one-dimensional (1-D) diffusion under a wide range of solution conditions. Our results have shown that the TALE search mechanism appears to be unique amongst the broad class of sequence-specific DNA binding proteins.
Recent advances in genome engineering offer the potential to dramatically alter the treatment of human disease. Achieving this potential, however, remains challenging due to the high degrees of precision and accuracy required for modifying large, intact genomes. Our group has used single molecule techniques to directly study the search process for transcription activator-like effector nucleases or TALE(N)s along DNA molecules, thereby revealing the molecular mechanisms underlying the DNA search process for these proteins. We use single molecule experiments to study TALE search along DNA based total internal reflection microscopy.Unexpectedly, our results show that TALEs utilize a two-state ‘search and check’ model for finding target sites while moving along DNA using a rotationally decoupled mechanism, despite remaining associated with DNA templates during the search process. In this way, TALE search is largely absent of rotationally coupled sliding. Our results suggest that the helical structure of TALEs enables these proteins to adopt a loose wrapped conformation around DNA templates during non-specific search, thereby facilitating rapid one-dimensional (1-D) diffusion under a wide range of solution conditions. Our results have shown that the TALE search mechanism appears to be unique amongst the broad class of sequence-specific DNA binding proteins.
Selected publications:
1. S. Jain, S. Shukla, C. Yang, M. Zhang, Z. Fatma, M. Lingamaneni, S. Abesteh, S. T. Lane, X. Xiong, Y. Wang, C. M. Schroeder, P. R. Selvin, H. Zhao, “TALEN Outperforms Cas9 in Editing Heterochromatin Target Sites”, Nature Communications, 12, 606 (2021).
2. L. Cuculis*, C. Zhao*, Z. Abil, H. Zhao, D. Shukla*, C. M. Schroeder*, “Divalent Cations Enhance TALE DNA-Binding Specificity”, Nucleic Acids Research, 48, 1406 (2020).
3. L. Cuculis, Z. Abil, H. Zhao, C. M. Schroeder, “TALE Proteins Search DNA using a Rotationally Decoupled Mechanism”, Nature Chemical Biology, 12, 831 (2016).
4. L. Cuculis, Z. Abil, H. Zhao, C. M. Schroeder, “Direct Observation of TALE Protein Dynamics Reveals a Two-state Search Mechanism”, Nature Communications, 6, 7277 (2015).
5. Y. Kim, S. Kim, J. A. Katzenellenbogen, C. M. Schroeder, “Specific Labeling of Zinc Finger Proteins using Noncanonical Amino Acids and Copper-free Click Chemistry”, Bioconjugate Chemistry, 23, 1891 (2012).
Flavin-binding fluorescent proteins (FbFPs) for anaerobic imaging
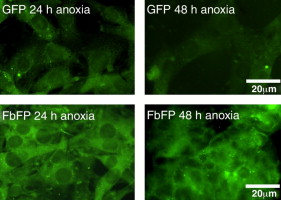 Recently, a new class of oxygen-independent fluorescent reporters was reported based on flavin-binding photosensors from Bacillus subtilis, Pseudomonas putida, and Arabidopsis thaliana. Flavin-binding fluorescent proteins (FbFPs) can function in the absence of oxygen, whereas the widely used green fluorescent protein (GFP) and related analogs strictly require oxygen for maturation of fluorescence. In our lab, we have applied the tools of directed evolution to isolate new and spectrally improved variants of FbFPs. Moving forward, we anticipate that spectrally enhanced FbFP variants will find pervasive use as reporter proteins for gene expression, subcellular localization and protein interactions in obligate and facultative anaerobes and in hypoxic niches of the human body (e.g. malignant tumors).
Recently, a new class of oxygen-independent fluorescent reporters was reported based on flavin-binding photosensors from Bacillus subtilis, Pseudomonas putida, and Arabidopsis thaliana. Flavin-binding fluorescent proteins (FbFPs) can function in the absence of oxygen, whereas the widely used green fluorescent protein (GFP) and related analogs strictly require oxygen for maturation of fluorescence. In our lab, we have applied the tools of directed evolution to isolate new and spectrally improved variants of FbFPs. Moving forward, we anticipate that spectrally enhanced FbFP variants will find pervasive use as reporter proteins for gene expression, subcellular localization and protein interactions in obligate and facultative anaerobes and in hypoxic niches of the human body (e.g. malignant tumors).
Selected publications:
1. A. Mukherjee, K. B. Weyant, J. Walker, U. Agrawal, I. Cann, C. M. Schroeder, “Engineering and Characterization of New LOV-based Fluorescent Proteins from Chlamydomonas reinhardtii and Vaucheria frigida“, ACS Synthetic Biology, 4, 371 (2015).
2. A. Mukherjee, J. Walker, K. B. Weyant, C. M. Schroeder, “Characterization of Flavin-based Fluorescent Proteins: An Emerging Class of Powerful Fluorescent Reporters”, PLOS ONE, 8, e64753 (2013).
3. A. Mukherjee, K. B. Weyant, J. Walker, C. M. Schroeder, “Directed Evolution of Bright Mutants of a Flavin-Dependent Anaerobic Fluorescent Protein from Pseudomonas putida“, Journal of Biological Engineering, 6, 20, (2012).
4. A. Mukherjee and C. M. Schroeder, “Flavin-based Fluorescent Proteins: Emerging Paradigms in Biological Imaging”, Current Opinion in Biotechnology, 31, 16 (2015).
Fluorescent dendrimer nanoconjugates
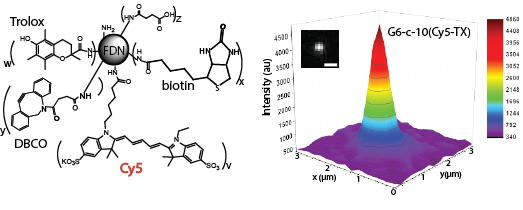 Advanced imaging techniques in the biological and chemical sciences critically rely on bright and photostable probes. We developed a new class of fluorescent molecules, fluorescent dendrimer nanoconjugates (FDNs), based on the covalent attachment of multiple fluorescent dyes onto a single dendritic scaffold. This results in extremely bright, nanometer scale, single-molecule probes that we have used for both traditional fluorescence microscopy, as well as for the burgeoning field of super-resolution microscopy where brightness is directly related to achievable resolution. In addition, we have engineered the photophysical properties of these probes by attaching the “photo-protective” triplet-state quencher Trolox directly onto the scaffold, an anti-fade agent normally used in solution. Trolox-conjugated probes are extremely photostable with vastly decreased amounts of transient dark states compared with single fluorescent dyes, and by modulating the amount of conjugated Trolox, we can exceed enhancement in photostability provided by adding large concentrations of Trolox in solution.
Advanced imaging techniques in the biological and chemical sciences critically rely on bright and photostable probes. We developed a new class of fluorescent molecules, fluorescent dendrimer nanoconjugates (FDNs), based on the covalent attachment of multiple fluorescent dyes onto a single dendritic scaffold. This results in extremely bright, nanometer scale, single-molecule probes that we have used for both traditional fluorescence microscopy, as well as for the burgeoning field of super-resolution microscopy where brightness is directly related to achievable resolution. In addition, we have engineered the photophysical properties of these probes by attaching the “photo-protective” triplet-state quencher Trolox directly onto the scaffold, an anti-fade agent normally used in solution. Trolox-conjugated probes are extremely photostable with vastly decreased amounts of transient dark states compared with single fluorescent dyes, and by modulating the amount of conjugated Trolox, we can exceed enhancement in photostability provided by adding large concentrations of Trolox in solution.
Selected publications:
1. D. T. Reilly, S. H. Kim, J. A. Katzenellenbogen, C. M. Schroeder, “Fluorescent Nanoconjugate Derivatives with Enhanced Photostability for Single Molecule Imaging”, Analytical Chemistry, 87, 11048 (2015).
2. Y. Kim, S. Kim, M. Tanyeri, J. A. Katzenellenbogen, C. M. Schroeder, Dendrimer Probes for Enhanced Photostability and Localization in Fluorescence Imaging, Biophysical Journal, 104, 1566 (2013).